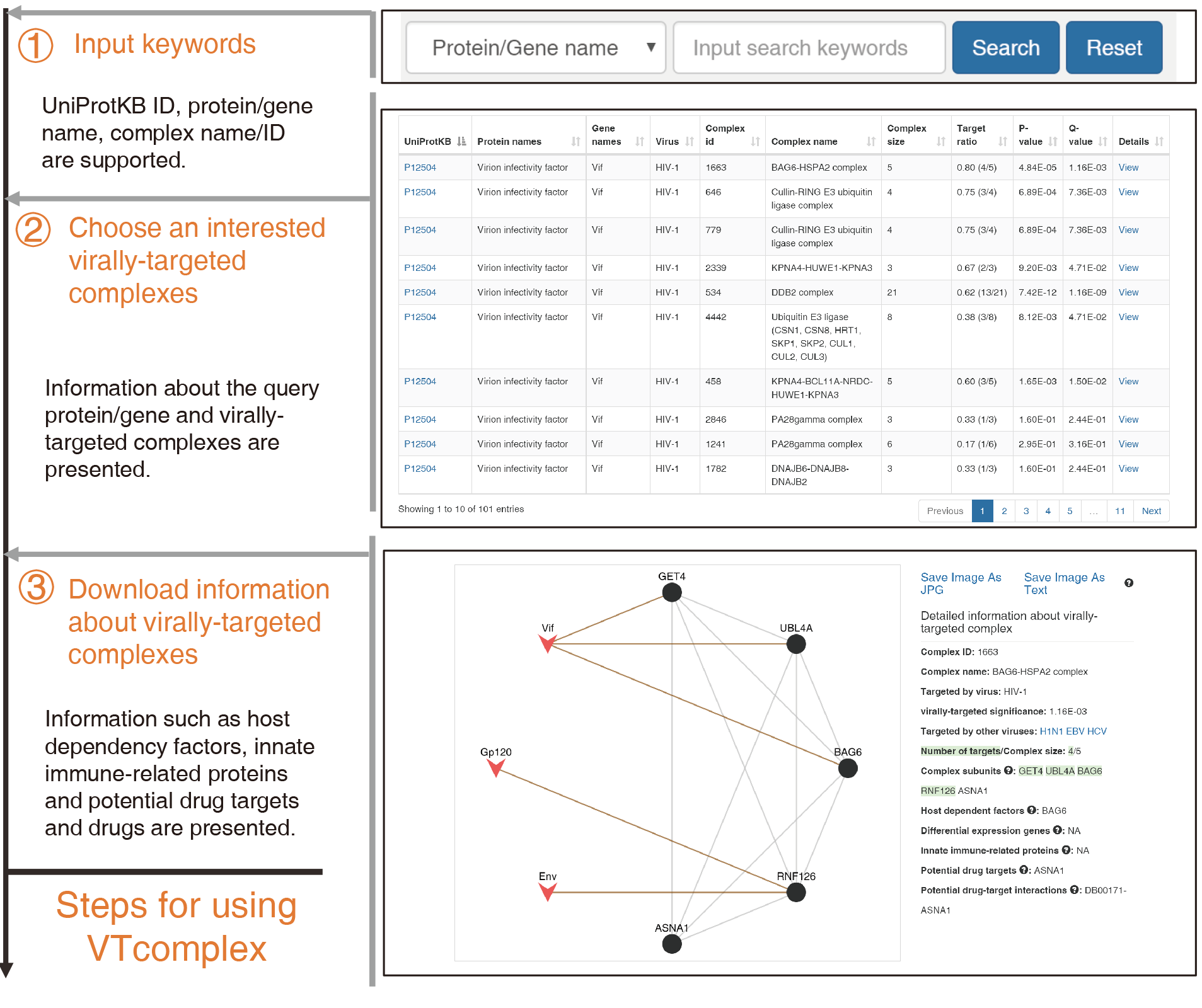
About VTcomplex
-
VTcomplex mainly contains virally-targeted human protein complexes and the associated information.
It effectively integrates several public resources such as
HPIDB, hu.MAP,
DrugBank, TTD
and DGIdb.
-
Currently, VTcomplex covers the data of five viruses including five viruses including influenza A virus subtype H1N1(H1N1),
human immunodeficiency virus subtype 1 (HIV-1), Epstein-Barr virus (EBV), human papillomavirus (HPV),
and hepatitis C virus (HCV).
User manual
-
Firstly, users can input searching keywords such as protein/gene name, UniProtKB ID, complex accession number
and complex name/ID. The searching result of virally-targeted complexes will be returned in table format.
-
Secondly, users can choose their interested virally-targeted complexes by complex name or VTsignificance and click the View for further view.
The VTsignificance is scoring variable used to determine the significance of a complex that was targeted by a specific virus.
Taking HIV-1 as an example, we first conduct Fisher’s exact test to calculate the significance of a complex (complex M) targeted by HIV-1.
The 2x2 contingency table is as follows:
| Within complex M | Outside complex M | |
|---|---|---|
| Targets | A | B |
| Non-targets | C | D |
A: the number of HIV-1’s targets within the complex M.
B: the number of HIV-1’s targets outside the complex M.
C: the number of HIV-1’s non-targets within the complex M.
D: the number of HIV-1’s non-targets outside the complex M.
The number HIV-1's targets: the total number of human proteins from human-HIV-1 PPIs.
The number HIV-1's non-targets: the total number of reviewed human proteins (20218) minus the total number of HIV-1's targets
The p-value of complex M is calculated by stats.fisher_exact([(A, B), (C, D)], alternative='greater') in scipy (https://docs.scipy.org)
The p-value obtained after Benjamini-Hochberg correction is defined as VTsignificance, which is used to determine the extent of the complex that was targeted by HIV-1.-
Thirdly, intraspecies and interspecies PPIs will be presented in network format on the left panel.
Other heterogeneous information within this complex, such as host dependent factors, innate immune-related genes,
dynamic information of subunits (differentially expressed genes, and periodic genes),
potential drug targets and drugs will be shown on the right panel. Currently, host dependent
factors and differentially expressed genes are only available for HIV-1