WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: ZLKZG1659504467 Submitted: 2022-08-03 13:27:47 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79747734 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh17G007370 (Len=545) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh17G007370, Len=545): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAGLTGIFDALIGFGGFGFFLSGVASICVFVGEMNGGNEVIAAPAAPPQPLDWKFSQVFGERTAGEEVQEVDIISAIEFD 80
WD40-rpts (_________________WD 1________________) (_________________WD
Query 81 KSGDHLATGDRGGRVVLFERTDTKDHGASRRDLERMDNAISRHPEFRYKTEFQSHEPEFDYLKSLEIEEKINKIRWCQPA 160
WD40-rpts 2_________________) (_________________WD 3________________) (______________
Query 161 NGALFLLSTNDKTIKYWKVQEKKVKKVSTMNVDPSKPVGNGSNASSSNSSSSGPYLANGGSPDRYPSNDPSFPARGIQSL 240
WD40-rpts WD 4_____________) (_________________WD 5_________________)
Query 241 RLPVVTTHETNLVARCRRVYGHAHDYHINSISNNSDGETFISADDLRINLWNLEISSQSFNIVDVKPANMEDLTEVITSA 320
WD40-rpts (________________WD 6_______________) (_______________
Query 321 EFHPTHCNMLAYSSSKGSIRLIDLRQSALCDSHAKLFEEQEAPGSRSFFTEIIASISDIKFGKEGRYILSRDYMTLKLWD 400
WD40-rpts __WD 7________________) (_________________WD 8_________________)
Query 401 INMDSGPVAAFQVHEYLRPKLCDLYENDSIFDKFECCLSGDGLRLATGSYSNLFRVFGCVSGSTEATTLEASKNPMRRQV 480
WD40-rpts (___________________WD 9__________________) (_________________
Query 481 QTPSRPARSLSSSITRVVRRGSESPGVDANGNSFDFTTKLLHLAWHPTENSIACAAANSLYMYYA 545
WD40-rpts _WD10_________________) (_________________WD11________________)
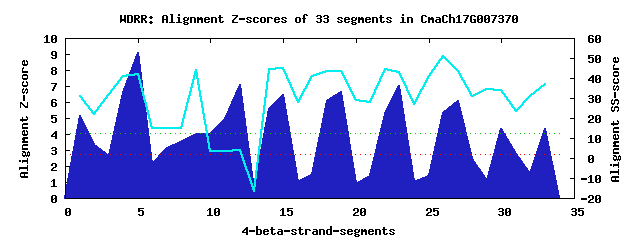 11 WD40-repeats found in QUERY=CmaCh17G007370 (Len=545): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 15-53 5.14 327.11 0.00243 17.5% 30.80 WD 2: Seg 5 60-99 9.06 488.91 1.6e-05 12.5% 42.02 WD 3: Seg 8 103-141 3.53 260.39 0.01912 2.5% 15.03 WD 4: Seg 9 146-178 3.99 279.31 0.01067 12.5% 44.03 WD 5: Seg 12 181-220 7.07 406.72 0.00021 7.5% 3.85 WD 6: Seg 15 256-292 6.42 379.84 0.00047 22.5% 45.19 WD 7: Seg 19 305-343 6.60 387.26 0.00038 25.0% 43.70 WD 8: Seg 23 361-400 6.98 402.82 0.00023 20.0% 42.94 WD 9: Seg 27 416-458 6.09 366.42 0.00072 22.5% 43.58 WD10: Seg 30 463-503 4.30 292.18 0.00717 12.5% 34.13 WD11: Seg 33 507-545 4.30 292.14 0.00718 5.0% 36.95Multiple sequence alignment of 11 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 15-53 -GG---FGFF-LSGVASICVFVGEMNG--G-N--EVIAAPAAPPQPLDW WD 2 60-99 GER---TAGE-EVQEVDIISAIEFDKS--G-D--HLATGDRGGRVVLFE WD 3 103-141 -TK---DHGA-SRRDLERMDNAISRHP--E-F--RYKTEFQSHEPEFDY WD 4 146-178 -------------EIEEKINKIRWCQP--A-NGALFLLSTNDKTIKYWK WD 5 181-220 EKK---VKKV-STMNVDPSKPVGNGSN--A-S--SSNSSSSGPYLANGG WD 6 256-292 CRR---VY---GHAHDYHINSISNNSD--G-E--TFISAD-DLRINLWN WD 7 305-343 --V---KPAN-MEDLTEVITSAEFHPT--HCN--MLAYSSSKGSIRLID WD 8 361-400 EAP---GSRSFFTEIIASISDIKFGKE--G-R--YILSRD-YMTLKLWD WD 9 416-458 YLRPKLCDLY-ENDSIFDKFECCLSGD--G-L--RLATGSYSNLFRVFG WD10 463-503 -ST---EATT-LEASKNPMRRQVQTPSRPA-R--SLSSSITRVVRRGSE WD11 507-545 -VD---ANGN-SFDFTTKLLHLAWHPT--E-N--SIACAAANSLYMYYA WD40 Template TGE---CLRT-LSGHTSGVTSVSFSPD--G-K--RLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |