WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: X5B0Q1659462168 Submitted: 2022-08-03 01:42:48 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79755302 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh07G007910 (Len=652) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh07G007910, Len=652): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MLSSDRDTDVFFDSEDYLPLEDSLVAEEVDSNKLGYEIWMSEPQSIKRRRQRFLQEVGLLEFASSRTCAHDLVIESSDSS 80
WD40-rpts (_________________WD 1_________________)
Query 81 GRMELERLTEFSGAVSSSDYAPTCNVEENDLELCSNEMLLPVSVSNSGSRRCHGEADVEGCRDIDVSKRKIKKWWRSFKN 160
WD40-rpts (_______________WD 2_______________) (________
Query 161 KAASLRRTEFSEELKTIPAESKTCRMKVYQNKKRCSEFSALYMRQQIVAHKGLIWTMKFSPDGKYLASGGEDGVVRVWRI 240
WD40-rpts _________WD 3________________) (_________________WD 4________________)
Query 241 SSANVSSEFLANDGNHNNKPREGKSSLSSKPLRFATVVIPEKVFQIDEQPVQKFYGHSSDVLDIAWSNSNCLLSSSKDKT 320
WD40-rpts (_________________WD 5________________) (________________WD 6____________
Query 321 VRLWGVGSDQCLGVFHHKNYVTCIQFNPMDENYFISGSIDGKIRVWSVLKQRVVDWADVRDVITALAYQPDGKGFAIGSI 400
WD40-rpts ____)(__________________WD 7__________________) (_________________WD 8_________
Query 401 TGTCRFYKTSGDYFHLDAQINVQGRKKASGKRITGIQFCRENFQRVMITSEDSKVRIFEGTEIVNKYKGLPKSGSQMSAS 480
WD40-rpts _______) (________________WD 9_______________) (_________________
Query 481 FTKNRKHIISVGEDSRVYMWNYDSLHLPSMKETKSQQSCEHFLSDGASVAIPWPGVGTELKCFGNNQRQFSQSQDCQGAA 560
WD40-rpts WD10________________) (_________________WD11________________) (________________
Query 561 ASWTRDSERFSLANWISADSLCRGSATWPEERLPLWDLPVADDEHGNLEHHNSQSDAHDHVAPPDTWGLVIVVAGLDGTI 640
WD40-rpts ________WD12________________________) (_________________WD13_____________
Query 641 KTFHNYGLPIRL 652
WD40-rpts ___)
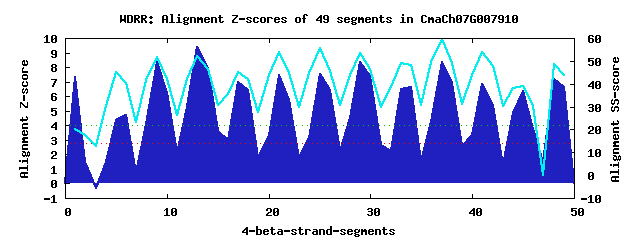 13 WD40-repeats found in QUERY=CmaCh07G007910 (Len=652): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-40 7.33 417.53 0.00015 7.5% 20.25 WD 2: Seg 6 113-148 4.74 310.49 0.00407 22.5% 40.45 WD 3: Seg 9 152-190 8.32 458.24 4.1e-05 17.5% 51.83 WD 4: Seg 13 201-239 9.35 500.98 1.1e-05 37.5% 52.20 WD 5: Seg 17 247-285 6.95 401.59 0.00024 20.0% 45.21 WD 6: Seg 21 288-325 7.43 421.58 0.00013 27.5% 53.71 WD 7: Seg 25 326-367 7.54 426.00 0.00011 37.5% 55.73 WD 8: Seg 29 370-408 8.33 458.59 4.1e-05 22.5% 53.52 WD 9: Seg 34 423-459 6.61 387.81 0.00037 17.5% 48.12 WD10: Seg 37 463-501 8.34 459.16 4.0e-05 20.0% 59.69 WD11: Seg 41 504-542 6.87 398.63 0.00026 12.5% 53.81 WD12: Seg 45 544-597 6.36 377.45 0.00051 12.5% 39.00 WD13: Seg 48 606-644 7.19 411.81 0.00018 22.5% 48.41Multiple sequence alignment of 13 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-40 ML-SSDRDTDVFFDSEDYLPLED-SL-VAEEVDS--------------NKL----GYEIWM WD 2 113-148 ---------LCSNEMLLPVSVSN-SG-SRRCHGE--------------ADVEGCRDIDVSK WD 3 152-190 -K-KWWRSFKNKAASLRRTEFSE-EL-KTIPAES--------------KTC----RMKVYQ WD 4 201-239 -L-YMRQQIVAHKGLIWTMKFSP-DG-KYLASGG--------------EDG----VVRVWR WD 5 247-285 -S-EFLANDGNHNNKPREGKSSL-SS-KPLRFAT--------------VVI----PEKVFQ WD 6 288-325 -E-QPVQKFYGHSSDVLDIAWS--NS-NCLLSSS--------------KDK----TVRLWG WD 7 326-367 VGSDQCLGVFHHKNYVTCIQFNPMDE-NYFISGS--------------IDG----KIRVWS WD 8 370-408 -K-QRVVDWADVRDVITALAYQP-DG-KGFAIGS--------------ITG----TCRFYK WD 9 423-459 QG-----RKKASGKRITGIQFCR-ENFQRVMITS--------------EDS----KVRIFE WD10 463-501 -I-VNKYKGLPKSGSQMSASFTK-NR-KHIISVG--------------EDS----RVYMWN WD11 504-542 -S-LHLPSMKETKSQQSCEHFLS-DG-ASVAIPW--------------PGV----GTELKC WD12 544-597 GN-NQRQFSQSQDCQGAAASWTR-DS-ERFSLANWISADSLCRGSATWPEE----RLPLWD WD13 606-644 -G-NLEHHNSQSDAHDHVAPPDT-WG-LVIVVAG--------------LDG----TIKTFH WD40 Template TG-ECLRTLSGHTSGVTSVSFSP-DG-KRLVSGS--------------EDG----TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |