WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: UQJG81659462786 Submitted: 2022-08-03 01:53:06 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79755895 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh09G005870 (Len=761) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh09G005870, Len=761): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MEDFAFHDGNVCGLLESRHINFRQDPTYSFPMHSSFIQRLSQEKELEGHQGCVNAVAWNSRGSLLISGSDDTRINIWSYS 80
WD40-rpts (_______________WD 1_______________) (_________________WD 2________________)(_
Query 81 GQKLLHSVETGHSANIFCTKFVPETSDELVVSGAGDAEVRLFNLSRLKGRRHDDSPIASSALYQCHTRRVKKLAVEIGNP 160
WD40-rpts ___________________WD 3___________________) (_________________WD 4__
Query 161 NVVWSASEDGTLRQHDFREGMSCSPEGASHQECHNVLLDLRCGAKRSLADPPRQTLALKSCDISSTRPHLLLVGGSDAFA 240
WD40-rpts _______________) (_________________WD 5_____________
Query 241 RLYDRRMLPPLSSSQKRMSPPPCVNYFCPMHLSERVRQGLHLTHVTFSPNGEEILLSYSGEHVYLMNVNHAGLGTMQYTS 320
WD40-rpts ___) (_________________WD 6_________________) (_________
Query 321 GDVSKLMSFTPILNGLELQHPVSNLSNGLPLNGGVNAKLDKCRKLLQIAEKCLEGGNYFGGIEACNEILDGFGRIIGVIL 400
WD40-rpts ________WD 7________________) (___________________WD 8__________________) (___
Query 401 KHDSLCSRAALLLKRKWKNDVHMAIRDCYSARKIDNSSFRAHYYMCEALSQLGRHKEALDFAFAAQCLAPSNPEVAEKVE 480
WD40-rpts ______________WD 9________________)
Query 481 SIKRDLAAAELEKSNKDNDGAIRSAPLGGVLSLSDLLHRSDANSDVSQDGVRSEREDSDYDDEVELDFETLSGDEAQDVD 560
WD40-rpts (________________WD10_______________) (_______________
Query 561 SNVLHGSVNLRIHRRVDPAIERVGANGSCGSPSSSHNDGILYQPEPVIDMKQRYVGHCNIGTDIKQASFLGQRGEYVASG 640
WD40-rpts __WD11_________________) (___________________WD12_________
Query 641 SDDGRWFIWEKKTGRLIKILVGDGAVVNCVQSHPFDCAIATSGIDNTIKLWTPTAPVRSVVAGGAIGPQAADVLAAIENN 720
WD40-rpts _________) (_________________WD13________________)(______________WD14_________
Query 721 QRRLCRNREAILPFEILERMHAEIGIVIVGGVAVSTPNILL 761
WD40-rpts _____)
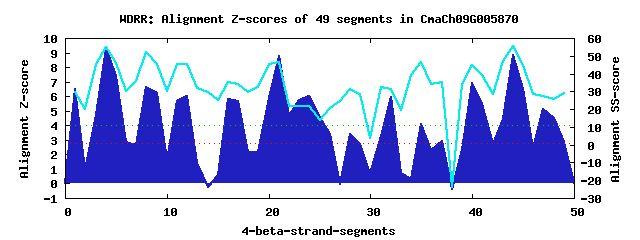 14 WD40-repeats found in QUERY=CmaCh09G005870 (Len=761): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-36 6.50 383.28 0.00043 7.5% 29.79 WD 2: Seg 4 40-78 9.33 500.13 1.1e-05 32.5% 55.08 WD 3: Seg 8 79-123 6.61 387.63 0.00037 20.0% 51.91 WD 4: Seg 12 137-176 6.00 362.71 0.00081 25.0% 45.25 WD 5: Seg 16 206-244 5.83 355.56 0.00100 12.5% 35.25 WD 6: Seg 21 268-307 8.78 477.41 2.3e-05 17.5% 46.24 WD 7: Seg 24 311-349 6.02 363.33 0.00079 10.0% 21.53 WD 8: Seg 26 353-395 3.41 255.36 0.02231 12.5% 20.68 WD 9: Seg 28 397-435 3.43 256.23 0.02172 10.0% 31.30 WD10: Seg 32 502-538 5.97 361.07 0.00085 10.0% 31.14 WD11: Seg 35 545-584 4.07 282.84 0.00957 17.5% 46.31 WD12: Seg 40 608-650 6.93 400.88 0.00025 25.0% 44.57 WD13: Seg 44 654-692 8.80 478.18 2.2e-05 27.5% 55.39 WD14: Seg 48 693-726 4.51 301.11 0.00544 10.0% 25.85Multiple sequence alignment of 14 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-36 -------MEDFAFHDGNVC-GL----LESRH--INF-RQDPTYSF----PMHSSF WD 2 40-78 -LS---QEKELEGHQGCVN-AV----AWNSR--GSL-LISGSDDT----RINIWS WD 3 79-123 YSGQKLLHSVETGHSANIF-CT----KFVPETSDEL-VVSGAGDA----EVRLFN WD 4 137-176 -IA---SSALYQCHTRRVK-KL----AVEIG--NPNVVWSASEDG----TLRQHD WD 5 206-244 -RS---LADPPRQTLALKS-CD----ISSTR--PHL-LLVGGSDA----FARLYD WD 6 268-307 CPM---HLSERVRQGLHLT-HV----TFSPN--GEE-ILLSYSGE----HVYLMN WD 7 311-349 -AG---LGTMQYTSGDVSK-LM----SFTPI--LNG-LELQHPVS----NLSNGL WD 8 353-395 -GG---VNAKLDKCRKLLQ-IA----EKCLE--GGN-YFGGIEACNEILDGFGRI WD 9 397-435 -GV---ILKHDSLCSRAAL-LL----KRKWK--NDV-HMAIRDCY----SARKID WD10 502-538 ------IRSAPLGGVLSLS-DL----LHRSD--ANS-DVSQDGVR----SEREDS WD11 545-584 -EL---DFETLSGDEAQDVDSN----VLHGS--VNL-RIHRRVDP----AIERVG WD12 608-650 -ID---MKQRYVGHCNIGT-DIKQASFLGQR--GEY-VASGSDDG----RWFIWE WD13 654-692 -GR---LIKILVGDGAVVN-CV----QSHPF--DCA-IATSGIDN----TIKLWT WD14 693-726 ---------PTAPVRSVVA-GG----AIGPQ--AAD-VLAAIENN----QRRLCR WD40 Template TGE---CLRTLSGHTSGVT-SV----SFSPD--GKR-LVSGSEDG----TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |