WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: TKSSK1348122591 Submitted: 2012-09-20 14:29:51 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 391128870 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = seq-TKSSK1348122591 (Len=1518) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (seq-TKSSK1348122591, Len=1518): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAAVGSGGYARNDAGEKLPSVMAGVPARRGQSSPPPAPPICLRRRTRLSTASEETVQNRVSLEKVLGITAQNSSGLTCDP 80
WD40-rpts (________________WD 1________________)_______________WD 2
Query 81 GTGHVAYLAGCVVVILDPKENKQQHIFNTARKSLSALAFSPDGKYIVTGENGHRPAVRIWDVEEKNQVAEMLGHKYGVAC 160
WD40-rpts ________________) (__________________WD 3_________________) (_______________
Query 161 VAFSPNMKHIVSMGYQHDMVLNVWDWKKDIVVASNKVSCRVIALSFSEDSSYFVTVGNRHVRFWFLEVSTETKVTSTVPL 240
WD40-rpts ___WD 4_________________) (________________WD 5________________) (____
Query 241 VGRSGILGELHNNIFCGVACGRGRMAGSTFCVSYSGLLCQFNEKRVLEKWINLKVSLSSCLCVSQELIFCGCTDGIVRIF 320
WD40-rpts ________________WD 6___________________) (_________________WD 7_________________
Query 321 QAHSLHYLANLPKPHYLGVDVAQGLEPSFLFHRKAEAVYPDTVALTFDPIHQWLSCVYKDHSIYIWDVKDINRVGKVWSE 400
WD40-rpts ) (____________________WD 8____________________)(____________
Query 401 LFHSSYVWNVEVYPEFEDQRACLPSGSFLTCSSDNTIRFWNLDSSPDSHWQKNIFSNTLLKVVYVENDIQHLQDMSHFPD 480
WD40-rpts ____________WD 9________________________)
Query 481 RGSENGTPMDVKAGVRVMQVSPDGQHLASGDRSGNLRIHELHFMDELVKVEAHDAEVLCLEYSKPETGLTLLASASRDRL 560
WD40-rpts (_______________WD10______________) (__________________WD11______________
Query 561 IHVLNVEKNYNLEQTLDDHSSSITAIKFAGNRDIQMISCGADKSIYFRSAQQGSDGLHFVRTHHVAEKTTLYDMDIDITQ 640
WD40-rpts ____) (_________________WD12________________)(_____________________WD13_____
Query 641 KYVAVACQDRNVRVYNTVNGKQKKCYKGSQGDEGSLLKVHVDPSGTFLATSCSDKSISVIDFYSGECIAKMFGHSEIITS 720
WD40-rpts _______________) (_________________WD14________________) (_______________
Query 721 MKFTYDCHHLITVSGDSCVFIWHLGPEITNCMKQHLLEIDHRQQQQHTNDKKRSGHPRQDTYVSTPSEIHSLSPGEQTED 800
WD40-rpts __WD15________________) (___________________WD16___________________)
Query 801 DLEEECEPEEMLKTPSKDSLDPDPRCLLTNGKLPLWAKRLLGDDDVADGLAFHAKRSYQPHGRWAERAGQEPLKTILDAQ 880
WD40-rpts (______________________WD17_____________________)
Query 881 DLDCYFTPMKPESLENSILDSLEPQSLASLLSESESPQEAGRGHPSFLPQQKESSEASELILYSLEAEVTVTGTDSQYCR 960
WD40-rpts (_________________WD18________________)
Query 961 KEVEAGPGDQQGDSYLRVSSDSPKDQSPPEDSGESEADLECSFAAIHSPAPPPDPAPRFATSLPHFPGCAGPTEDELSLP 1040
WD40-rpts
Query 1041 EGPSVPSSSLPQTPEQEKFLRHHFETLTESPCRALGDVEASEAEDHFFNPRLSISTQFLSSLQKASRFTHTFPPRATQCL 1120
WD40-rpts
Query 1121 VKSPEVKLMDRGGSQPRAGTGYASPDRTHVLAAGKAEETLEAWRPPPPCLTSLASCVPASSVLPTDRNLPTPTSAPTPGL 1200
WD40-rpts
Query 1201 AQGVHAPSTCSYMEATASSRARISRSISLGDSEGPIVATLAQPLRRPSSVGELASLGQELQAITTATTPSLDSEGQEPAL 1280
WD40-rpts
Query 1281 RSWGNHEARANLRLTLSSACDGLLQPPVDTQPGVTVPAVSFPAPSPVEESALRLHGSAFRPSLPAPESPGLPAHPSNPQL 1360
WD40-rpts
Query 1361 PEARPGIPGGTASLLEPTSGALGLLQGSPARWSEPWVPVEALPPSPLELSRVGNILHRLQTTFQEALDLYRVLVSSGQVD 1440
WD40-rpts
Query 1441 TGQQQARTELVSTFLWIHSQLEAECLVGTSVAPAQALPSPGPPSPPTLYPLASPDLQALLEHYSELLVQAVRRKARGH 1518
WD40-rpts
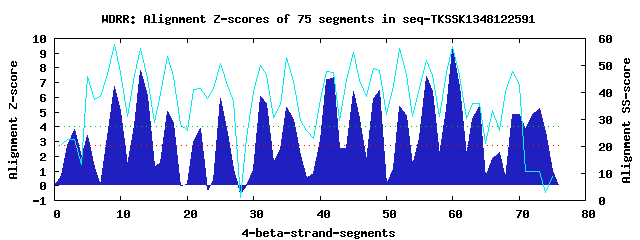 18 WD40-repeats found in QUERY=seq-TKSSK1348122591 (Len=1518): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 3 24-61 3.83 272.71 0.01308 15.0% 22.71 WD 2: Seg 5 60-97 3.39 254.88 0.02265 7.5% 45.80 WD 3: Seg 9 101-141 6.71 391.83 0.00033 25.0% 57.85 WD 4: Seg 13 145-185 7.80 436.81 8.1e-05 37.5% 56.27 WD 5: Seg 17 188-225 5.07 324.13 0.00266 17.5% 53.39 WD 6: Seg 22 236-280 3.88 274.87 0.01224 12.5% 41.58 WD 7: Seg 25 282-321 5.90 358.31 0.00092 10.0% 50.76 WD 8: Seg 31 342-387 6.04 364.10 0.00077 20.0% 49.88 WD 9: Seg 35 388-441 5.30 333.57 0.00199 25.0% 52.80 WD10: Seg 42 486-520 7.33 417.51 0.00015 27.5% 47.40 WD11: Seg 45 524-565 6.37 377.72 0.00051 25.0% 54.87 WD12: Seg 49 571-609 6.46 381.71 0.00045 22.5% 48.26 WD13: Seg 52 610-656 5.35 335.72 0.00186 7.5% 56.41 WD14: Seg 56 663-701 7.41 420.78 0.00013 25.0% 51.97 WD15: Seg 60 705-743 9.26 497.25 1.2e-05 32.5% 57.20 WD16: Seg 64 753-796 5.36 335.94 0.00185 15.0% 35.88 WD17: Seg 69 830-878 4.84 314.71 0.00357 12.5% 47.62 WD18: Seg 73 882-920 5.25 331.55 0.00212 17.5% 10.66Multiple sequence alignment of 18 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 24-61 -GV-------P-----A---------RRGQS-----SPPPAPPICLRRRT-------------RLSTAS--EE--TVQNRVS WD 2 60-97 -VS-------L-----E---------KVLGI-----TAQNSSGLTCDPGT---G---------HVAYLA---G--CVVVILD WD 3 101-141 -NK-------Q-----Q---------HIFNT-----ARKSLSALAFSPDG---K---------YIVTGE--NGHRPAVRIWD WD 4 145-185 -KN-------Q-----V---------AEMLG-----HKYGVACVAFSPNM---K---------HIVSMGYQHD--MVLNVWD WD 5 188-225 -KD-------I-----V---------VASNK-----VSCRVIALSFSEDS---S---------YFVTVG---N--RHVRFWF WD 6 236-280 STV-------P-----L---------VGRSGILGELHNNIFCGVACGRGR---M---------AGSTFC--VS--YSGLLCQ WD 7 282-321 NEK-------R-----V---------LEKWI-----NLKVSLSSCLCVSQ---E---------LIFCGC--TD--GIVRIFQ WD 8 342-387 AQG-LEPSFLF-----H---------RKAEA-----VYPDTVALTFDPIH---Q---------WLSCVY--KD--HSIYIWD WD 9 388-441 VKD-------INRVGKV---------WSELF-----HSSYVWNVEVYPEF---EDQRACLPSGSFLTCS--SD--NTIRFWN WD10 486-520 --------------------------GTPMD-----VKAGVRVMQVSPDG---Q---------HLASGD--RS--GNLRIHE WD11 524-565 -MD-------E-----L---------VKVEA-----HDAEVLCLEYSKPETGLT---------LLASAS--RD--RLIHVLN WD12 571-609 --N-------L-----E---------QTLDD-----HSSSITAIKFAGNR---D--------IQMISCG--AD--KSIYFRS WD13 610-656 AQQGSDGLHFV-----R---------THHVA-----EKTTLYDMDIDITQ---K---------YVAVAC--QD--RNVRVYN WD14 663-701 -KK-------C-----Y---------KGSQG-----DEGSLLKVHVDPSG---T---------FLATSC--SD--KSISVID WD15 705-743 -GE-------C-----I---------AKMFG-----HSEIITSMKFTYDC---H---------HLITVS--GD--SCVFIWH WD16 753-796 KQH-------L-----L-----EIDHRQQQQ-----HTNDKKRSGHPRQD---T---------YVSTPS--EI--HSLSPGE WD17 830-878 NGK-------L-----PLWAKRLLGDDDVAD-----GLAFHAKRSYQPHG---R---------WAERAG--QE--PLKTILD WD18 882-920 -LD-------C-----Y---------FTPMK-----PESLENSILDSLEP---Q---------SLASLL--SE--SESPQEA WD40 Template TGE-------C-----L---------RTLSG-----HTSGVTSVSFSPDG---K---------RLVSGS--ED--GTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |