WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: OB6SE1659497825 Submitted: 2022-08-03 11:37:05 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79752879 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh10G003230 (Len=949) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh10G003230, Len=949): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MSSLPRIFLLFSLCFSLFPSSFSLSHPDPLTIRPFQVNHSPPATIPAFPEQSEVQGCRLDLPDDLFHGIKTACGPSKGGV 80
WD40-rpts
Query 81 SGNLHRSRCCPVLAAWLYAAYSATALRRPVRASAGPGHTAPSYDLPLLPDDSETCVSNLDQALNQKGIELMKPNETCDVV 160
WD40-rpts
Query 161 YCYCGIRLHPLSCPEAFSLTHDGILEGDRNVKRLERNCLSSSNSNGFPGLGGCSKCLSTLYQLNNKEALNSSKPENRTIK 240
WD40-rpts
Query 241 MHHTECQLMGLTWLLAKNRTAYIRTVTSVLRAKMMSKDGSDPRSCTLNSDGMPLAVDSAEISGSSVSVSIQAPFSQIDSP 320
WD40-rpts (_______________________WD 1_______________________)(____________
Query 321 FRHFQHLLFFFFFFIALIMATAELCETYACVPSTERGRGILISGHPKTNSVLYTNGRSVIILNLDNPLEVSVYAEHAYPA 400
WD40-rpts ________WD 2___________________)(_____________WD 3____________) (_____________
Query 401 TVARYSPNGEWIASADVSGTVRIWGTHSGFVLKKEFKVLTGRIDDLQWSPDGMRIVVCGEGKGKSLVRAFMWDSGTNVGE 480
WD40-rpts ____WD 4________________)(____________________WD 5____________________) (_____
Query 481 FDGHSRRVLSCGFKPTRPFRIATCGEDFLVNFYEGPPFRFKLSLRNHSNFVNCARFSPDGSKFISVSSDKKGIIYDAKTG 560
WD40-rpts ____________WD 6_________________) (_________________WD 7________________) (
Query 561 DKMGELSPDDGHKGSIYAVSWSSDGKQVLTVSADKTAKVWVISDDGNGKLVKTLTSPGTGGIDDMLVGCLWQNQHLVTVS 640
WD40-rpts __________________WD 8__________________) (__________________WD 9__________
Query 641 LGGTISLFSANDLEKTPVILSGHMKNVTSLVVLKSDPKVILSTSYDGLIIKWIQGIGFSGKLQRRENSQIKCFAALEDEL 720
WD40-rpts ________) (_________________WD10_________________)(_________________WD11_____
Query 721 VTSGFDNKVWRVSIKNGECGEAEAIDVGSQPKDLTLAALSPELALVSIDSGVVLLRGSKVVSTVNLGFTVTASVLAPDGS 800
WD40-rpts ____________) (_______________WD12_______________)(_________________WD13__
Query 801 EAIIGGEDGKLHIYSVVGDSLAEEATLERHRGAVSVIRYSPDLSMFASGDLNREAIVWDRASREVKLKNMLYHTARINCL 880
WD40-rpts ______________) (_________________WD14________________)(___________________W
Query 881 AWSPDNTKVATGSLDTCIIIYEIDKPASNRLTIKGAHLGGVYGLAFTDDYSVVSSGEDACVRVWKMVPQ 949
WD40-rpts D15__________________) (________________WD16________________)
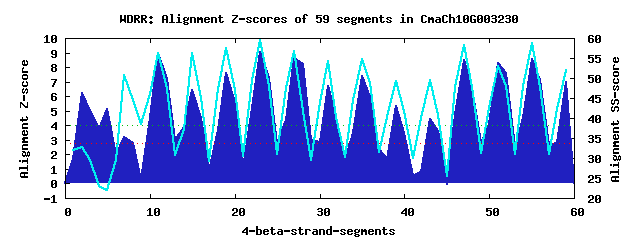 16 WD40-repeats found in QUERY=CmaCh10G003230 (Len=949): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 256-307 6.21 371.38 0.00062 22.5% 32.71 WD 2: Seg 5 308-352 5.13 326.49 0.00248 7.5% 21.90 WD 3: Seg 8 353-383 2.78 229.46 0.04918 7.5% 44.49 WD 4: Seg 11 387-425 8.94 483.93 1.9e-05 22.5% 56.19 WD 5: Seg 15 426-471 6.40 378.95 0.00049 22.5% 56.26 WD 6: Seg 19 475-514 7.59 428.36 0.00010 25.0% 57.43 WD 7: Seg 23 518-556 9.04 488.04 1.6e-05 30.0% 59.62 WD 8: Seg 27 560-601 8.61 470.36 2.8e-05 42.5% 56.79 WD 9: Seg 31 608-649 6.70 391.51 0.00033 27.5% 54.35 WD10: Seg 35 654-693 7.40 420.33 0.00013 40.0% 54.74 WD11: Seg 39 694-733 5.33 334.71 0.00192 12.5% 49.34 WD12: Seg 43 741-776 4.45 298.56 0.00588 15.0% 49.55 WD13: Seg 47 777-815 8.52 466.52 3.2e-05 25.0% 58.30 WD14: Seg 51 821-859 8.29 457.07 4.3e-05 30.0% 52.96 WD15: Seg 55 860-902 8.58 469.28 2.9e-05 25.0% 58.87 WD16: Seg 59 908-945 6.98 403.02 0.00023 30.0% 51.92Multiple sequence alignment of 16 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 256-307 AK-----NRTAYIRTVTSVLRAKMMS---KDGSDPRS---CTLNSDGM-PLAVDSAE---ISGSSVS WD 2 308-352 VSIQAPFS---QIDSP---------F---RHFQHLLF---FFFFFIAL-IMATAELC---ETYACVP WD 3 353-383 -------------------------S---TERGRGIL---ISGHPKTN-SVLY-TNG---RSVIILN WD 4 387-425 -P-----L---EVSVY---------A---EHAYPATV---ARYSPNGE-WIASADVS---GTVRIWG WD 5 426-471 TH-----SGFVLKKEF---------K---VLTGRIDD---LQWSPDGM-RIVVCGEGKGKSLVRAFM WD 6 475-514 -G-----T---NVGEF---------D---GHSRRVLS---CGFKPTRPFRIATCGED---FLVNFYE WD 7 518-556 -F-----R---FKLSL---------R---NHSNFVNC---ARFSPDGS-KFISVSSD---KKGIIYD WD 8 560-601 -G-----D---KMGEL---------SPDDGHKGSIYA---VSWSSDGK-QVLTVSAD---KTAKVWV WD 9 608-649 -G-----K---LVKTL---------T---SPGTGGIDDMLVGCLWQNQ-HLVTVSLG---GTISLFS WD10 654-693 -E-----K---TPVIL---------S---GHMKNVTS--LVVLKSDPK-VILSTSYD---GLIIKWI WD11 694-733 QG-----I---GFSGK---------L---QRRENSQI---KCFAALED-ELVTSGFD---NKVWRVS WD12 741-776 -E-----A---EAIDV---------G---SQPKDLTL---AALSPE----LALVSID---SGVVLLR WD13 777-815 -G-----S---KVVST---------V---NLGFTVTA---SVLAPDGS-EAIIGGED---GKLHIYS WD14 821-859 -L-----A---EEATL---------E---RHRGAVSV---IRYSPDLS-MFASGDLN---REAIVWD WD15 860-902 RA-----SREVKLKNM---------L---YHTARINC---LAWSPDNT-KVATGSLD---TCIIIYE WD16 908-945 -S-----N---RLTIK---------G---AHLGGVYG---LAFTDDYS--VVSSGED---ACVRVWK WD40 Template TG-----E---CLRTL---------S---GHTSGVTS---VSFSPDGK-RLVSGSED---GTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |