WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: MR4IJ1659500163 Submitted: 2022-08-03 12:16:03 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79750036 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh12G003120 (Len=910) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh12G003120, Len=910): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MSQTNWEADKMLDVYIHDYLVKRDLKATAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFWDIFIARTNEKHSDVAASYIE 80
WD40-rpts
Query 81 TQLIKAREQQQHQQHQQQQQQQQPQAQQQQPQHMQMLLMQRHAQQQQQQQQQQQQQQQQQQQQQSQQQQQQQTQQQQQPR 160
WD40-rpts
Query 161 RDGAQLLNGSSNGFVGNDPLMRQNPGSVNALATKMYEDRLKLPLQRDSLDEAAMKQRYGENVGQLLDPNHASILKSAAAT 240
WD40-rpts
Query 241 SQSSGQVLHSTAGGMSPQVQSRSQQLAGSAPMQDIKSEINAGLNPRAAGPEGSLMGIPGSNHGGNNLTLKGWPLTGLEQL 320
WD40-rpts
Query 321 RSGILQQQKSFIQAPQQFPQLQMLTPQHQQLMLAQQNLTSPSVNDDGRRLRMLLNSRISKDGLSNSVGDVVPNVGSPLQA 400
WD40-rpts
Query 401 GSPLLQRGDNTEMMLKIKIAQLQHQQQHQQNNSQQQQQQQQLQQHALSNPQSQSSNHNLHQQEKIGGAGSVTMDGSMSNS 480
WD40-rpts (_________________WD 1____________
Query 481 FRGNDQVSKNQTGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSTHTPGDAISMPALPHSGSSSKPLIMFSSDGTGT 560
WD40-rpts ____) (__________________WD 2________
Query 561 FTSPSNQLWDDKELELQADMDRFVEDGSLDDNVESFLSHDDTDPRDPVGRCMDGSTGFTFTEVNSVRASSSKVTSCHFSS 640
WD40-rpts _________) (________________WD 3________________) (_________________WD
Query 641 DGKLLASGGHDKKAVLWYTENLKPKTSLEEHGSMITDVRFSPSMPRLATSSFDRTVRVWDADNRCYSLRSFTGHSTSVMS 720
WD40-rpts 4________________) (_________________WD 5________________) (_______________
Query 721 LDFHPKKDDLIGSCDGDGEIRYWNITNGSCAAVFKGGTGQLRFQPRLGRYLGAAVDNVVSIFDVETQARLHTLQGHTKTI 800
WD40-rpts __WD 6_________________) (_______________WD 7_______________) (_____________
Query 801 NSLCWDPSGEFLASVSEDSVRVWSLASGSEGESIHELSINGNKFHSCVFHPTFSTLLVIGCYQSLEFWNTSENKTMTLSA 880
WD40-rpts ___WD 8________________) (_________________WD 9________________)(__________
Query 881 HEGLISALAVSTASGLVASASHDKFIKLWK 910
WD40-rpts ________WD10_________________)
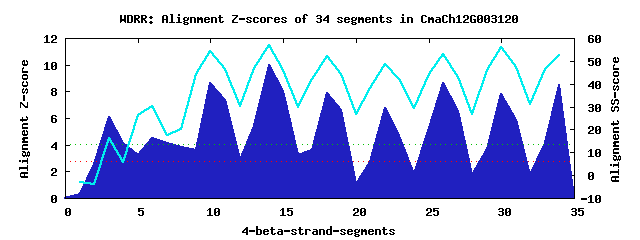 10 WD40-repeats found in QUERY=CmaCh12G003120 (Len=910): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 3 447-485 6.08 365.71 0.00073 15.0% 16.34 WD 2: Seg 6 530-570 4.48 299.73 0.00568 20.0% 30.17 WD 3: Seg 8 576-613 3.84 273.13 0.01291 12.5% 20.27 WD 4: Seg 10 620-658 8.66 472.58 2.7e-05 35.0% 54.28 WD 5: Seg 14 662-700 10.01 528.18 4.7e-06 40.0% 57.00 WD 6: Seg 18 705-744 7.86 439.18 7.5e-05 32.5% 52.01 WD 7: Seg 22 748-783 6.78 394.87 0.00030 22.5% 48.43 WD 8: Seg 26 787-824 8.66 472.57 2.7e-05 40.0% 52.92 WD 9: Seg 30 831-869 7.80 436.98 8.0e-05 22.5% 56.19 WD10: Seg 34 870-910 8.51 466.08 3.3e-05 32.5% 52.37Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 447-485 LSN-PQSQS--S----NHNL-HQQEKIGGAGS-VTMD----GSMSNSFRGND WD 2 530-570 TPG-DAISM--PALPHSGSSSKPLIMFSSDGT-GTFT----SPSN---QLWD WD 3 576-613 ---------LQA----DMDRFVEDGSLDDNVE-SFLSHDDTDPRDPVGRCMD WD 4 620-658 -FT-EVNSV--R----ASSSKVTSCHFSSDGK-LLAS----GGHDKKAVLWY WD 5 662-700 -LK-PKTSL--E----EHGSMITDVRFSPSMP-RLAT----SSFDRTVRVWD WD 6 705-744 -CY-SLRSF--T----GHSTSVMSLDFHPKKDDLIGS----CDGDGEIRYWN WD 7 748-783 -GS-CAAVF--K----GGTG---QLRFQPRLG-RYLG----AAVDNVVSIFD WD 8 787-824 -QA-RLHTL--Q----GHTKTINSLCWDPSGE-FLAS----VSED-SVRVWS WD 9 831-869 -GE-SIHEL--S----INGNKFHSCVFHPTFS-TLLV----IGCYQSLEFWN WD10 870-910 TSENKTMTL--S----AHEGLISALAVSTASG-LVAS----ASHDKFIKLWK WD40 Template TGE-CLRTL--S----GHTSGVTSVSFSPDGK-RLVS----GSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |