WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: IWU221659462159 Submitted: 2022-08-03 01:42:39 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79788726 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh07G004300 (Len=535) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh07G004300, Len=535): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 SSQFPYGGFSGGATATMDGEHGGERLVVQGRAEIDTRAPFKSVKEAVTLFGERILVGEIYAKKMKERVEVGESQAKQNEE 80
WD40-rpts (_________________________________________________________WD 1_
Query 81 NNLLAFCLQSLKDELERTKQELEQLKSTEHEKPHHEHPLSSTMAIHPDVDKDPKFVQNKKENNTNMLQKKRSVKFAVPLE 160
WD40-rpts ________________________________________________________)
Query 161 LNQNISVKYESMAQKSSLSPSSTKKPKKKTLAYIWKRRAKFSSMGGAQSEREEKVSLELTEETLQSMEVGMTFRDYNGRI 240
WD40-rpts (_______________________WD 2______________________) (_____________
Query 241 SSMDFHRTSSYLVTASDDESIRLYDVGSATCLKTINSKKYGVDLVCFTSHPTTVIYSSKNGWDESLRLLSLNDNKYLRYF 320
WD40-rpts ____WD 3________________) (__________________WD 4__________________) (______
Query 321 KGHHDRVVSLSLCARKECFISGSLDQTVLLWDQRAEKCQGLLRVQGKPTTAYDEQGVVFAIAYGGYIRMFDSRKYEKGPF 400
WD40-rpts ___________WD 5________________)(_________________WD 6________________) (_
Query 401 DIFSVGEDVSDTNVVKFSNDGRLMLLTTSDGHIHVLDSFRGTLLSTYNVKPISSNSTLEASFSPEGMFVISGSGDGSVYA 480
WD40-rpts ________________WD 7________________) (__________________WD 8_________________
Query 481 WSVRSGKEVASWMSSEMEPPVIKWAPGSLMFATGSSELSFWIPDLSKLGAYVDRK 535
WD40-rpts _) (_________________WD 9________________)
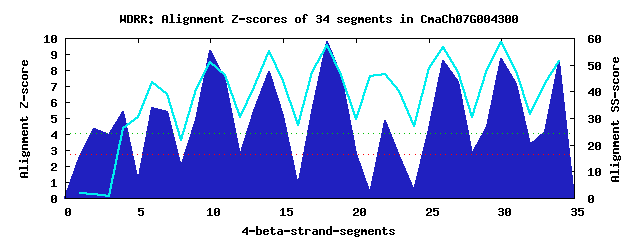 9 WD40-repeats found in QUERY=CmaCh07G004300 (Len=535): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 18-137 4.33 293.58 0.00686 10.0% 1.52 WD 2: Seg 6 171-221 5.60 345.82 0.00136 15.0% 43.68 WD 3: Seg 10 227-265 9.18 493.80 1.4e-05 25.0% 51.12 WD 4: Seg 14 269-310 7.86 439.24 7.5e-05 22.5% 55.22 WD 5: Seg 18 314-352 9.78 518.64 6.4e-06 35.0% 57.37 WD 6: Seg 22 353-391 4.83 314.30 0.00361 12.5% 46.53 WD 7: Seg 26 399-437 8.56 468.22 3.0e-05 27.5% 56.60 WD 8: Seg 30 441-482 8.66 472.29 2.7e-05 35.0% 58.33 WD 9: Seg 34 486-524 8.52 466.45 3.2e-05 12.5% 51.43Multiple sequence alignment of 9 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 18-137 DGEHGGERLVVQGRAEIDTRAPFKSVKEAVTLFGERILVGEIYAKKMKERVEVGESQAKQNEENNLLAFCLQSLKDELERTKQELEQLK-----------ST---EHEKPHHEHPLSSTMAIHPDV---DKDPKFVQ WD 2 171-221 SMA---------------QKSS--------------L---------------------------------------------------SPSSTKKPKKKTLA---YIWKRRAKFSSMGGAQSEREE---KVSLELTE WD 3 227-265 -ME---------------VGMT--------------F---------------------------------------------------R-----------DY---NGRISSMDFHRTSSYLVTASD---DESIRLYD WD 4 269-310 -AT---------------CLKT--------------I---------------------------------------------------N-----------SK---KYGVDLVCFTSHPTTVIYSSKNGWDESLRLLS WD 5 314-352 -NK---------------YLRY--------------F---------------------------------------------------K-----------GH---HDRVVSLSLCARKECFISGSL---DQTVLLWD WD 6 353-391 QRA---------------EKCQ--------------G---------------------------------------------------L-----------LR---VQGKPTTAYDEQGV-VFAIAY---GGYIRMFD WD 7 399-437 -PF---------------DIFS--------------V---------------------------------------------------G-----------ED---VSDTNVVKFSNDGRLMLLTTS---DGHIHVLD WD 8 441-482 -GT---------------LLST--------------Y---------------------------------------------------N-----------VKPISSNSTLEASFSPEGMFVISGSG---DGSVYAWS WD 9 486-524 -GK---------------EVAS--------------W---------------------------------------------------M-----------SS---EMEPPVIKWAPGSLMFATGSS---ELSFWIPD WD40 Template TGE---------------CLRT--------------L---------------------------------------------------S-----------GH---TSGVTSVSFSPDGKRLVSGSE---DGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |