WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: GUPHR1659514028 Submitted: 2022-08-03 16:07:08 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79734007 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CsaV3_1G031710 (Len=810) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CsaV3_1G031710, Len=810): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MITGGKSYVSAPPAFSNDAKRLLVCTGTSVSIFSTSTGLQIASLKGHKAFVTSVTVVPASSAASKILCFCWTTSLDGTIR 80
WD40-rpts (______________WD 1_____________) (____________________WD 2__________________
Query 81 YWDFSIPELMKTIDIRLPVYSMVIPSLLGQLLERDVKSRDLFAYVSVQNIGVKDGKPVPVRGQILKCNLTKSRLATGVIL 160
WD40-rpts __) (_________________WD 3________________) (_______
Query 161 AETQQPEYLTTSSSGSFFGIRNKRKIHVWKVPNGQFEKLGAKKITLHHTKDLTVLAFHPTQRTVAAGDVTGRILIWRGFG 240
WD40-rpts _________WD 4________________) (_________________WD 5________________)(__
Query 241 NRTFPVSGEEAGKKSFDSDEDRPGVRGNDDADSCSTRHWHPTEVIALSFSSDGAYLYSGGKEGVLVVWQLDTEKRKYLPR 320
WD40-rpts ___________WD 6____________) (_________________WD 7________________) (________
Query 321 IGSPLLYFTDSPDPLLASVSCADNQIHLLKMPSMEILKSISGIKLPCSFPDVCQGSNNGFAFNQNDGLVALRSENYSIQF 400
WD40-rpts _________WD 8________________) (__________________WD 9________________
Query 401 YSLFDDCGICEVQICERNHQPGEELTVVITSVVLSLDGSLMTTAEIRIPEGGIGGLICLKFWDSELENKKFSLSTVVYEP 480
WD40-rpts _) (_____________________WD10_____________________)(________________
Query 481 HRDAGISALAFHPNRRMVVSTSYGGDFKIWVCNGGLPKVQGEKNSSWMCHSVGSYKKKSMTAATFSADGSVLAVAAETVI 560
WD40-rpts _____WD11_____________________) (________________WD12_____________
Query 561 TLWDPEQNILVAVIGETLTPIVNLSFAGDSQFLVSVSQGSKPQLSVWTVSKLSISWSYKLHIEALACAVDMSSFAVLALI 640
WD40-rpts ___) (__________________WD13_________________) (_________________WD14_______
Query 641 PESVRLQFSDSTFQGRDGMILHFNANDPVPLSTWSVRKAQGGGLAFLRSEKSNISSDEKSGHPWLVYINGDHEYTLFDPS 720
WD40-rpts _________) (_________________WD
Query 721 GKEGQELSLTKQGSYHALEETGGKFGYEAIYGELPEFVSKMDQTLSAPSVPSQRPWETIFSGSSHELPPLTKLCSAFLES 800
WD40-rpts 15________________)
Query 801 LLERRTVTTE 810
WD40-rpts
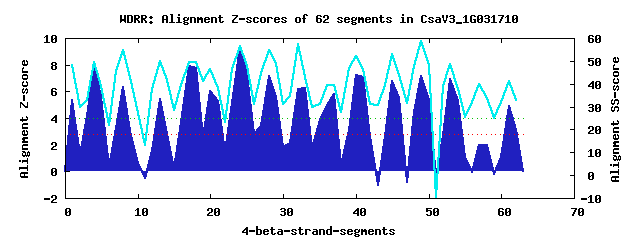 15 WD40-repeats found in QUERY=CsaV3_1G031710 (Len=810): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 2-34 5.32 334.34 0.00194 17.5% 48.03 WD 2: Seg 4 38-83 7.95 443.17 6.6e-05 40.0% 49.69 WD 3: Seg 8 87-125 6.35 377.09 0.00052 15.0% 54.73 WD 4: Seg 13 153-190 5.44 339.36 0.00166 15.0% 50.08 WD 5: Seg 17 199-237 7.90 441.19 7.0e-05 22.5% 49.69 WD 6: Seg 21 238-268 5.31 334.05 0.00196 17.5% 38.26 WD 7: Seg 24 271-309 9.30 498.63 1.2e-05 35.0% 56.62 WD 8: Seg 28 312-350 7.18 411.32 0.00018 20.0% 54.83 WD 9: Seg 33 362-402 6.24 372.60 0.00059 10.0% 42.78 WD10: Seg 37 416-463 5.83 355.56 0.00101 25.0% 39.40 WD11: Seg 40 464-511 7.25 414.02 0.00016 25.0% 52.09 WD12: Seg 45 527-564 6.80 395.39 0.00029 27.5% 53.08 WD13: Seg 49 568-608 7.15 410.15 0.00018 22.5% 58.70 WD14: Seg 53 612-650 6.91 400.20 0.00025 10.0% 48.64 WD15: Seg 61 701-739 4.93 318.30 0.00319 12.5% 41.03Multiple sequence alignment of 15 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 2-34 -------ITG--------GKSYVSAPPAFSNDAKRLL-------VCT-GT---------S--VSIFS WD 2 38-83 -GLQ-IASLK--------GHKAFVTSVTVVPASSAASKILCFCWTTSLDG---------T--IRYWD WD 3 87-125 -PEL-MKTID--------IRLPVYSMVIPSLLGQLLE-------RDVKSR---------D--LFAYV WD 4 153-190 -RLA-TGVIL--------AETQQPEYLTTSSSGSFFG-------IRN-KR---------K--IHVWK WD 5 199-237 -LGA-KKITL--------HHTKDLTVLAFHPTQRTVA-------AGDVTG---------R--ILIWR WD 6 238-268 ------------------GFGNRTFPVSGEEAGKKSF-------DSDEDR---------P--GVRGN WD 7 271-309 -ADS-CSTRH--------WHPTEVIALSFSSDGAYLY-------SGGKEG---------V--LVVWQ WD 8 312-350 -TEK-RKYLP--------RIGSPLLYFTDSPDPLLAS-------VSCADN---------Q--IHLLK WD 9 362-402 GIKLPCSFPD--------VCQGSNNGFAFNQNDGLVA-------LRSENY---------S--IQFYS WD10 416-463 -ERN-HQPGE--------ELTVVITSVVLSLDGSLMT-------TAEIRIPEGGIGGLIC--LKFWD WD11 464-511 SELE-NKKFSLSTVVYEPHRDAGISALAFHPNRRMVV-------STSYGG---------D--FKIWV WD12 527-564 -WMC-HSVGS--------YKKKSMTAATFSADGSVLA-------VAA-ET---------V--ITLWD WD13 568-608 -NIL-VAVIG--------ETLTPIVNLSFAGDSQFLV-------SVSQGS---------KPQLSVWT WD14 612-650 -LSI-SWSYK--------LHIEALACAVDMSSFAVLA-------LIPESV---------R--LQFSD WD15 701-739 -GHP-WLVYI--------NGDHEYTLFDPSGKEGQEL-------SLTKQG---------S--YHALE WD40 Template TGEC-LRTLS--------GHTSGVTSVSFSPDGKRLV-------SGSEDG---------T--IKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |