WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: GKS0C1659462346 Submitted: 2022-08-03 01:45:46 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79788149 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh08G012500 (Len=625) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh08G012500, Len=625): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MASSNIRDLLTSFSPDLHFFAISSGDGRIKIWDTLKGQIQTEFADFFTTDSTSILTKSGKGHLSVDYKCMKWLSLEKKRK 80
WD40-rpts (______________WD 1_____________)(______________WD 2______________)(____________
Query 81 RKRQCSLLLLGTGSGDVMALDVAAGELKWQISDCHPGGVATISFPTHGSRIYTAGADGMLCEIDSMTGSLLRKFKASTKA 160
WD40-rpts __WD 3______________)(___________________WD 4__________________) (____________
Query 161 ISCISVSPDGKKIATAASQMKIFNCSDHKKIQKFSGHPGAVRCMVFTEDGKYILSSGVGERYIVVWSLGGEKKQSASCVL 240
WD40-rpts ____WD 5_______________) (_________________WD 6_________________)(____________
Query 241 AMEHPAIFMDSRRSGGGGVDETALYVLAISEIGVCYLWYGQNLEELRSVKPTKVLISNNDISSKSKKRVTPSIYAAQFQG 320
WD40-rpts _____WD 7_________________) (_______________________WD 8__________
Query 321 IPKSGSGQVFLAHGLLVKPSFQNVVVHSGTDINLNSSNDGILLPSHSVGKSKKGLDVQGGVVALDRANAEDALCPIPKVF 400
WD40-rpts _____________) (_________________WD 9________________) (_________________WD1
Query 401 DSQEKRTLYHDLHVDRDDVMTELVDSGSQVEDVVGVEDSAAVCLEDKLRLLGILDSTDDHTSESILKSAIFKGIDLEANM 480
WD40-rpts 0________________)
Query 481 SQKKLREAVLSLAPGDACKLLGNLVSIWQSRLRSGKNVLPWIYSLLLNHNQHILSQEQSTQILDSLFKFTKSKETAVQPL 560
WD40-rpts
Query 561 LQLSGRLQLVLAQIERASANKTDQTVRHGPEIDGGESSNDEDEEVHDVLYGEEEYESELSSDNEN 625
WD40-rpts
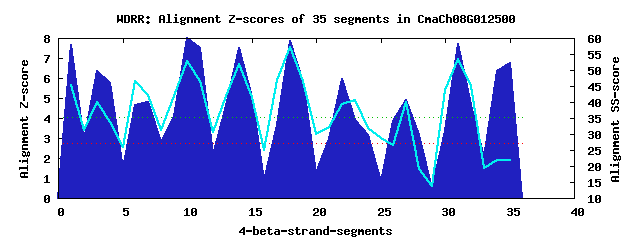 10 WD40-repeats found in QUERY=CmaCh08G012500 (Len=625): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-33 7.66 430.96 9.7e-05 30.0% 45.31 WD 2: Seg 4 34-67 5.74 351.67 0.00113 20.0% 33.64 WD 3: Seg 7 68-101 4.79 312.71 0.00380 12.5% 42.17 WD 4: Seg 10 102-144 8.00 445.10 6.2e-05 25.0% 52.94 WD 5: Seg 14 148-184 7.49 424.21 0.00012 27.5% 51.95 WD 6: Seg 18 188-227 7.85 439.05 7.5e-05 35.0% 57.09 WD 7: Seg 22 228-267 5.96 360.90 0.00085 15.0% 39.36 WD 8: Seg 27 283-334 4.89 316.86 0.00334 17.5% 39.85 WD 9: Seg 31 338-376 7.72 433.44 9.0e-05 22.5% 53.53 WD10: Seg 35 380-418 6.76 393.84 0.00031 10.0% 21.82Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-33 ----------MASSNIRDLLTSFSPDLHF-FAISS-------------GDGRIKIWD WD 2 34-67 ---------TLKGQIQTEFADFFTTDSTS-ILTKS-------------GKGHLSVDY WD 3 68-101 KCM---KWLSLEKKRK------RKRQCSL-LLLGT-------------GSGDVMALD WD 4 102-144 VAAGELKWQISDCHPGGVATISFPTHGSR-IYTAG-------------ADGMLCEID WD 5 148-184 -GS---LLRKFKASTKAISCISVSPDGKK-IATAA-------------SQ--MKIFN WD 6 188-227 -HK---KIQKFSGHPGAVRCMVFTEDGKYILSSGV-------------GERYIVVWS WD 7 228-267 LGG---EKKQSASCVLAMEHPAIFMDSRR-SGGGG-------------VDETALYVL WD 8 283-334 -LE---ELRSVKPTKVLISNNDISSKSKK-RVTPSIYAAQFQGIPKSGSGQVFLAHG WD 9 338-376 -KP---SFQNVVVHSGTDINLNSSNDGIL-LPSHS-------------VGKSKKGLD WD10 380-418 -GV---VALDRANAEDALCPIPKVFDSQE-KRTLY-------------HDLHVDRDD WD40 Template TGE---CLRTLSGHTSGVTSVSFSPDGKR-LVSGS-------------EDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |