WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: G7E2B1659459578 Submitted: 2022-08-03 00:59:38 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79791385 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh01G002640 (Len=1382) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh01G002640, Len=1382): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAVEEEQTEWHLHSGQYLGEISALCFLHLPPQISSLPILLAGSGSEVLAYNLESGNMIESFRVFEGIRVHGISSISLNFS 80
WD40-rpts (_________________WD 1_________________) (___________________
Query 81 EASSCTKLDFVLVVFGEKRVKLYRVSVEMVAEVCVHLVPLCSLPRFNHWVLDACFLKSRDSSSSAGSDSCGYIAIGCSDN 160
WD40-rpts WD 2___________________) (______________________WD 3_________________
Query 161 SVHVWDTCESRMILEVESPESCLLYSMRLWGDAIETLRVASGTIFNEIIVWEVVPSEGTLKDCDEKSNDIQFDHLKYEAM 240
WD40-rpts _____) (___________________WD 4__________________) (_
Query 241 HISRLVGHEGSIFRITWSSDGSKLVSVSDDRSARIWRLNAKRSEADKPEDVIVLFGHNARVWDCCIYNSLIITAGEDCTC 320
WD40-rpts ________________WD 5________________) (________________WD 6____________
Query 321 RAWGLDGKQLEMIKEHIGRGVWRCLYDPMSSLLITAGFDSSIKVHQLNISLSGTSNDPAEITSSSMKRELFTSCIPKSLD 400
WD40-rpts ___) (_________________WD 7________________)________________WD 8______________
Query 401 HNGLMDSKSEYVRCLRFSSEFTLYVATNHGYLYHATLSDAMDVKWTKVIHINEEVPIVCMDLFPSSPSRVSCEADDWVAL 480
WD40-rpts __)(______________WD 9______________)(____________________WD10__________________
Query 481 GDGQGRMTVLKVLESSNAPKLHLSFNWSAEAERQLLGTFWCKSLGFRYIFTADPRGVLKLWRLNDHVPASQYGENCNVSL 560
WD40-rpts _) (______________________WD11_____________________) (_
Query 561 IAKYVSCFGLRIMCVDASFEEEIVVCGDVRGNLILFPLSKDLSLDNPITTEVKIIPTCYFKGAHGISTVTSVVIRLDSCQ 640
WD40-rpts ________________WD12________________) (_______________WD1
Query 641 TEIHSTGADGCICHMEYVKLKDQKLLEFIGMKQVRELTSVQSLFYDQNSFLDLTSNLYAIGFASTDFIIWNLITESKVLQ 720
WD40-rpts 3______________) (_______________________WD14_______________________) (__
Query 721 IQCGGWRRPYSHYLGYVPELKNCFAYVKDEIIYIHRYWVSDGGKKIFPQNLHVQFHGRELHSVRFIPEGVEPKADNKHDI 800
WD40-rpts _______________WD15________________) (________________________WD16____
Query 801 LSRSSWIVTGCEDGTVRLTRYNPSTNNWSASNLLGEHVGGSAVRSICYISEVHMISSDGTIMPDVKDIQESDPDLTENPV 880
WD40-rpts ___________________) (________________WD17________________)(______________
Query 881 LLISAGAKRVLTSWLQKHRKLEKMEGTSACLRQNEEVRNEPSRLASSISFKWLSTDMPTKRSSSHRNSYNIREDEAKSSY 960
WD40-rpts __WD18________________) (______
Query 961 SINPVAESNLLQEKEELSLKSCPVEKYEDDWRYLAVTGFLVKHFNSRITVCFIVVACSDATLSLRALILPNRLWFDVASL 1040
WD40-rpts ____________WD19__________________) (__
Query 1041 VPVGSPVLTLQHVVVPKFHPNGAGQTLFGNVYIVISGATDGSIAFWDLTGNIEAFMKRLSSLHQAMFIDFQKRPRTGRGS 1120
WD40-rpts ____________________WD20______________________)
Query 1121 QGGRWRRSLSTVTQGQSSKDLITKKDGDNTILSIQNQVPCKGSSKADISEADTVCSEPVCCASSELVLTSGNSSSKMSEI 1200
WD40-rpts (___________________WD21___________________)(
Query 1201 RPIHVLTNVHQSGVNCLHVAAVNSPECVNNCFLYHVISGGDDQALQCLTFDLSFLSESPISEIMESESGSMKRFILHSED 1280
WD40-rpts _______________WD22_______________) (_________________WD23________________)
Query 1281 CNRKYMVRFLRSLKIASAHSSAIKGVWTDGIWVFSTGLDQRIRCWKLEAQGKLVEYTYLIITVPEPEAIDARACDKNHYQ 1360
WD40-rpts (_______________WD24______________)
Query 1361 IAVAGRGMQVIVFSTSLDTGRR 1382
WD40-rpts
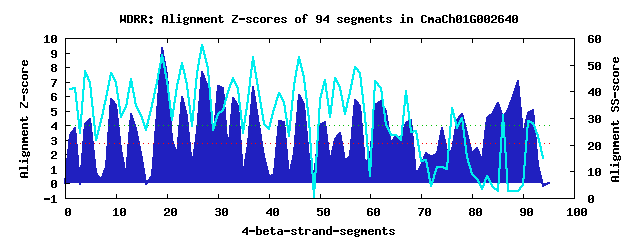 24 WD40-repeats found in QUERY=CmaCh01G002640 (Len=1382): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 12-51 3.81 272.04 0.01336 7.5% 41.08 WD 2: Seg 5 61-104 4.46 298.71 0.00586 12.5% 43.16 WD 3: Seg 9 117-166 5.83 355.64 0.00100 25.0% 47.03 WD 4: Seg 13 170-212 4.80 313.05 0.00376 17.5% 44.64 WD 5: Seg 19 239-277 9.31 499.18 1.2e-05 30.0% 53.63 WD 6: Seg 23 288-324 5.96 360.99 0.00085 22.5% 50.66 WD 7: Seg 27 328-366 7.65 430.61 9.8e-05 20.0% 57.21 WD 8: Seg 30 365-403 6.68 390.54 0.00034 12.5% 31.67 WD 9: Seg 34 404-437 5.33 334.85 0.00191 10.0% 41.02 WD10: Seg 37 438-482 6.62 388.21 0.00036 12.5% 52.71 WD11: Seg 42 494-542 4.32 293.09 0.00697 15.0% 39.47 WD12: Seg 46 559-597 6.11 367.14 0.00070 15.0% 52.73 WD13: Seg 51 622-656 4.24 289.82 0.00771 12.5% 44.08 WD14: Seg 54 660-711 3.44 256.60 0.02148 10.0% 41.13 WD15: Seg 57 718-756 5.75 352.19 0.00112 10.0% 49.17 WD16: Seg 62 768-820 5.69 349.75 0.00120 30.0% 41.28 WD17: Seg 65 828-865 3.31 251.51 0.02511 25.0% 23.79 WD18: Seg 68 866-903 4.36 294.92 0.00659 10.0% 24.60 WD19: Seg 74 954-995 3.80 271.64 0.01352 12.5% 11.77 WD20: Seg 78 1038-1087 4.77 311.64 0.00392 25.0% 30.18 WD21: Seg 85 1156-1199 5.55 343.86 0.00144 17.5% 2.73 WD22: Seg 86 1200-1235 4.47 299.16 0.00577 12.5% 31.11 WD23: Seg 89 1239-1277 7.05 405.80 0.00021 20.0% 2.49 WD24: Seg 92 1292-1326 5.08 324.51 0.00263 27.5% 28.21Multiple sequence alignment of 24 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 12-51 LH---S-------------GQY---------L----GEIS----A--L-----------CFLHLPPQ------I---------------S-----SLPILLAGSGSEVLAY---N WD 2 61-104 -F---R-----VF------EGI---------R----VHGI----S--S-----------ISLNFSEA------S----------SCTKLD-------FVLVVFGEKRVKLY---R WD 3 117-166 -L---V-----PL------CSL---------P----RFNH----W--VLDACFLKSRDSSSSAGSDS------C---------------G-------YIAIGCSDNSVHVW---D WD 4 170-212 -S---R-----MI------LEV---------E----SPESCLLYS--M-----------RLWGDAIE------T---------------L-------RVASGTIFNEIIVW---E WD 5 239-277 -A---M-----HI------SRL---------V----GHEG----S--I-----------FRITWSSD------G---------------S-------KLVSVSDDRSARIW---R WD 6 288-324 -P---E-----DV------IVL---------F----GHNA----R--V-----------WDCCIY--------N---------------S-------LIITAGEDCTCRAW---G WD 7 328-366 -K---Q-----LE------MIK---------E----HIGR----G--V-----------WRCLYDPM------S---------------S-------LLITAGFDSSIKVH---Q WD 8 365-403 -H---Q-----LN------ISL---------S----GTSN----D--P-----------AEITSSSM------K---------------R-------ELFTSCIPKSLDHN---G WD 9 404-437 --------------------LM---------D----SKSE----Y--V-----------RCLRFSSE------F---------------T-------LYVATNHGYLYHAT---L WD10 438-482 SD---AMDVKWTK------VIH---------I----NEEV----P--I-----------VCMDLFPS------S---------------P-------SRVSCEADDWVALG---D WD11 494-542 ES---S-----NA------PKLHLSFNWSAEA----ERQL----L--G-----------TFWCKSLG------F---------------R-------YIFTADPRGVLKLW---R WD12 559-597 -S---L-----IA------KYV---------S----CFGL----R--I-----------MCVDASFE------E---------------E-------IVVCGDVRGNLILF---P WD13 622-656 GA---H-----GI------STV---------T---------------S-----------VVIRLDSC------Q---------------T-------EIHSTGADGCICHM---E WD14 660-711 LK---D-----QKLLEFIGMKQ---------V----RELT----S--V-----------QSLFYDQNSFLDLTS---------------N-------LYAIGFASTDFIIW---N WD15 718-756 -V---L-----QI------QCG---------G----WRRP----Y--S-----------HYLGYVPE------L---------------K-------NCFAYVKDEIIYIH---R WD16 768-820 PQ---N-----L--------HV---------Q----FHGR----E--L-----------HSVRFIPE------GVEPKADNKHDILSRSS-------WIVTGCEDGTVRLT---R WD17 828-865 -W---S-----AS------NLL---------G----EHVG----GSAV-----------RSICYISE------V---------------H-------MISS---DGTIMPD---V WD18 866-903 -----K-----DI------QES---------D----PDLT----E--N-----------PVLLISAG------A---------------K-------RVLTSWLQKHRKLE---K WD19 954-995 -D---E-----AK------SSY---------S----INPV----A--E-----------SNLLQEKE------E---------------L-------SLKSCPVEKYEDDWRYLA WD20 1038-1087 ASLVPV-----GS------PVL---------T----LQHV----V--V-----------PKFHPNGA------G---------------QTLFGNVYIVISGATDGSIAFW---D WD21 1156-1199 NQ---V-----PC------KGS---------SKADISEAD----T--V-----------CSEPVCCA------S---------------S-------ELVLTSGNSSSKMS---E WD22 1200-1235 IR---P-----IH------VLT---------N----VHQS----G--V-----------NCLHVAA--------------------------------VNSPECVNNCFLY---H WD23 1239-1277 -G---G-----DD------QAL---------Q----CLTF----D--L-----------SFLSESPI------S---------------E-------IMESESGSMKRFIL---H WD24 1292-1326 -----------SL------KIA---------S----AHSS----A--I-----------KGV--WTD------G---------------I-------WVFSTGLDQRIRCW---K WD40 Template TG---E-----CL------RTL---------S----GHTS----G--V-----------TSVSFSPD------G---------------K-------RLVSGSEDGTIKVW---D |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |