WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: G5EOV1659500056 Submitted: 2022-08-03 12:14:16 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79751274 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh11G017850 (Len=854) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh11G017850, Len=854): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MPGEAALSAIVDSTNLKASGFDDELHSQVDFINLLKRTIESLKLRSCHRPPSVLLRCCHGQQRLQKREKSVGGLTHTLLL 80
WD40-rpts (____________WD 1____________) (________________WD 2_______________)
Query 81 YQVNPLLPPLRPFQASMETNSNDYKLRCELMGHEDDVRGICVCGNVGIATSSRDRTVRFWNLDGRKYEQSKIFLGHTSFV 160
WD40-rpts (________________WD 3_______________) (_____________
Query 161 GPLAWISPDEQYPEGRIVSGGMDTFVIVWDLRTGERVQTLKGHQQQVTGIALDSGDIVSSSVDCTLRRWRNGQVVEFWGA 240
WD40-rpts ______WD 4___________________) (________________WD 5_______________)(_________
Query 241 HNAAIQSVIKLPSGELVTGSSDATLKLWRGKSCLKTFSGHTDTVRSLSVMSDLGVLSASHDGSIRLWALSGQTLMEMVGH 320
WD40-rpts ________WD 6________________)(_________________WD 7________________) (_________
Query 321 ASIVYSVDSHASGLIVSGSEDCSAKIWKDGICVQSIEHPGCVWDAKFLENGDIVTACSDGVLRVWTADQERIAEPQELES 400
WD40-rpts _______WD 8________________)(________________WD 9________________)
Query 401 FASRLSQYKLKSKRVGGLKLEELPGMEALQIPGTSNGQTKVIREGDNGVAYSWNSKDYKWDKIGEVVDGPDDSRGKPVLD 480
WD40-rpts (_____________________WD10___________
Query 481 GAEYDFVFDVDIGDGEPIRKLPYNLSDDPYTTADNWLLKENLPLVYRQQVVDFILQNSGKTNFVVDPSFRDPYTGSSAYV 560
WD40-rpts __________)
Query 561 PGGPSNVSDESHKPVFKHIPKKGALVFDVAQFDGILKKIVEFNNALLADSDKKNYSLPEIEVSRLGALVKILKDTSHYHS 640
WD40-rpts
Query 641 TKFTDADVALLLNLLRTWPCELLFPVIDTLRMTVLHPDGAIMLLKLVDSDDILLELIQKVTTPPAFGANLLTSIRLIANL 720
WD40-rpts
Query 721 FKNSGYYDWLQKRRSEIIDAYSSCYSAANKAVQLSFSTLILNFAVLLIEKKDFDGQCQVLSAAIECKISEESLEADSKFR 800
WD40-rpts
Query 801 ALVAIGSLMIEGGDDMTRTALDFDVKSIARKASASKDAKIAEVGADIELLIKDR 854
WD40-rpts
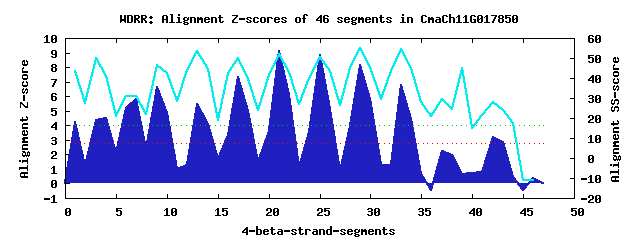 10 WD40-repeats found in QUERY=CmaCh11G017850 (Len=854): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-30 4.20 288.31 0.00808 10.0% 43.51 WD 2: Seg 4 32-68 4.51 300.76 0.00550 12.5% 40.96 WD 3: Seg 9 105-141 6.61 387.55 0.00037 22.5% 46.33 WD 4: Seg 13 147-190 5.49 341.32 0.00156 35.0% 53.40 WD 5: Seg 17 194-230 7.29 415.59 0.00016 37.5% 50.00 WD 6: Seg 21 231-269 9.08 489.58 1.6e-05 35.0% 52.22 WD 7: Seg 25 270-308 8.84 479.66 2.1e-05 42.5% 50.40 WD 8: Seg 29 311-348 8.17 452.19 5.0e-05 42.5% 54.93 WD 9: Seg 33 349-386 6.79 395.10 0.00029 27.5% 54.71 WD10: Seg 42 444-491 3.20 246.78 0.02902 20.0% 28.07Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-30 ------------------MPGEAALSAIVD-----STNLKASGFDDELHSQVD WD 2 32-68 INLLKRT-----------IESLKLRSCHRP-----PSVLLRCCHGQQRLQKRE WD 3 105-141 KLRC--ELM--------GHEDDVRGICVCG-----NVG-IATSSRDRTVRFWN WD 4 147-190 -YEQSKIFL--------GHTSFVGPLAWISPDEQYPEGRIVSGGMDTFVIVWD WD 5 194-230 -GERVQTLK--------GHQQQVTGIAL-------DSGDIVSSSVDCTLRRWR WD 6 231-269 NGQVVEFWG--------AHNAAIQSVIKLP-----SGE-LVTGSSDATLKLWR WD 7 270-308 GKSCLKTFS--------GHTDTVRSLSVMS-----DLG-VLSASHDGSIRLWA WD 8 311-348 -GQTLMEMV--------GHASIVYSVDSHA-----SG-LIVSGSEDCSAKIWK WD 9 349-386 DGICVQSI---------EHPGCVWDAKFLE-----NGD-IVTACSDGVLRVWT WD10 444-491 EGDNGVAYSWNSKDYKWDKIGEVVDGPDDS-----RGKPVLDGAEYDFVFDVD WD40 Template TGECLRTLS--------GHTSGVTSVSFSP-----DGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |