WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: CBMM01487439940 Submitted: 2017-02-19 01:45:40 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 251811786 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ambra1-trimmed (Len=712) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ambra1-trimmed, Len=712): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MKVVPEKNAVRILWGRERGARAMGAQRLLQELVEDKTRWMKWEGKRVELPDSPRSTFLLAFSPDRTLLASTHVNHNIYIT 80
WD40-rpts (_______________WD 1_______________)(_________________WD 2________________
Query 81 EVKTGKCVHSLIGHRRTPWCVTFHPTISGLIASGCLDGEVRIWDLHGGSESWFTDSNNAIASLAFHPTAQLLLIATANEI 160
WD40-rpts ) (_________________WD 3_________________) (________________WD 4_____________
Query 161 HFWDWSRREPFAVVKTASEMERVRLVRFDPLGHYLLTAIVNPSNQQGARMSAPSLGRFVPRRFLLPEYLPYAGIFHERGQ 240
WD40-rpts ___)(_____________________WD 5_____________________) (
Query 241 PGLATHSSVNRVLAGAVIGDGQSAVASNIANTTYRLQWWDFTKFDLPEISNASVNVLVQNCKIYNDASCDISADGQLLAA 320
WD40-rpts _________________WD 6________________) (_____________________WD 7___________
Query 321 FIPSSQRGFPDEGILAVYSLAPHNLGEMLYTKRFGPNAISVSLSPMGRYVMVGLASRRILLHPSTEHMVAQVFRLQQAHG 400
WD40-rpts __________) (___________________WD 8__________________)
Query 401 GETSMRRVFNVLYPMPADQRRHVSINSARWLPEPGLGLAYGTNKGDLVICRPEALNSGVEYYWDQLNETVFTVHSNSRSS 480
WD40-rpts (_________________WD 9_______
Query 481 ERPGTSRATWRTDRDMGLMNAIGLQPRNPATSVTSQGTQTLALQLQNAETQTEREVPEPGTAASGPGEGEGSEYGASGED 560
WD40-rpts __________) (_______________WD10______________)
Query 561 ALSRIQRLMAEGGMTAVVQREQSTTMASMGGFGNNIIVSHRIHRSSQTGTEPGAAHTSSPQPSTSRGLLPEAGQLAERGL 640
WD40-rpts
Query 641 SPRTASWDQPGTPGREPTQPTLPSSSPVPIPVSLPSAEGPTLHCELTNNNHLLDGGSSRGDAAGPRGEPRNR 712
WD40-rpts
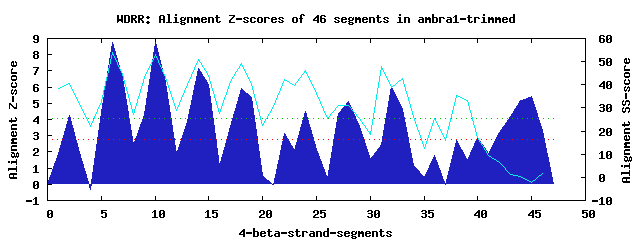 10 WD40-repeats found in QUERY=ambra1-trimmed (Len=712): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 7-42 4.26 290.83 0.00747 20.0% 40.57 WD 2: Seg 6 43-81 8.75 476.11 2.4e-05 22.5% 53.75 WD 3: Seg 10 85-124 8.77 477.07 2.3e-05 35.0% 53.12 WD 4: Seg 14 127-164 7.15 410.13 0.00018 20.0% 50.84 WD 5: Seg 18 165-212 5.91 358.59 0.00091 17.5% 49.21 WD 6: Seg 24 240-278 4.52 301.25 0.00541 12.5% 45.99 WD 7: Seg 28 284-331 5.11 325.67 0.00254 15.0% 30.85 WD 8: Seg 32 341-383 6.01 362.72 0.00080 22.5% 38.98 WD 9: Seg 42 452-491 3.20 247.05 0.02879 7.5% 6.22 WD10: Seg 45 493-527 5.40 337.77 0.00175 7.5% -2.36Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 7-42 -KN----AVRI---LWGRERGA------------RAMGA-QRLLQEL----VEDKTRWMKW WD 2 43-81 EGK----RV-E---LPDSPRST---------FLLAFSPD-RTLLAST----HVNHNIYITE WD 3 85-124 -GK----CVHS---LIGHRRTP---------WCVTFHPTISGLIASG----CLDGEVRIWD WD 4 127-164 -GG----SESW---FTDSNNAI---------ASLAFHPT-AQLLLIA----TAN-EIHFWD WD 5 165-212 WSRREPFAVVK---TASEMERV---------RLVRFDPL-GHYLLTAIVNPSNQQGARMSA WD 6 240-278 -QP----GLAT---HSSVNRVL---------AGAVIGDG-QSAVASN----IANTTYRLQW WD 7 284-331 -FD----LPEI---SNASVNVLVQNCKIYNDASCDISAD-GQLLAAF----IPSSQRGFPD WD 8 341-383 APH----NLGEMLYTKRFGPNA---------ISVSLSPM-GRYVMVG----LASRRILLHP WD 9 452-491 PEA----LNSG---VEYYWDQL---------NETVFTVH-SNSRSSE----RPGTSRATWR WD10 493-527 ---------DR---DMGLMNAI---------GLQPRNPA-TSVTSQG----TQTLALQLQN WD40 Template TGE----CLRT---LSGHTSGV---------TSVSFSPD-GKRLVSG----SEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |