WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: AJ80X1463471386 Submitted: 2016-05-17 15:49:46 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 275781103 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = tr_F1RA29_F1RA29_DANRE (Len=1843) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (tr_F1RA29_F1RA29_DANRE, Len=1843): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MSSKALWQQPYVNIFKHMKIEEWKKVSKEGDVSLYMDKNLKCSVFSIRGSIQASNYILLPKTSSQTLGLTGRYLYLLFKP 80
WD40-rpts
Query 81 SPNKHFIVHLDIAAEEGQVIRVSFSNLFKEFKSTATWLQFPFLCGAAHGSVDENTSTTAKQGLVGPAPWSTRWTCLTMDL 160
WD40-rpts
Query 161 RYVLSVYLNRTHSHLKSIKLCANMSVKNIITSDLLFDPGLSFSEFRQSGVMLPDGSAPMPREMCFPVPKGQSWHHLYDYI 240
WD40-rpts
Query 241 RFPSDGIKSPIDFIQKASNSAAKKASAEKESPQREESRMVRISKPVNDRVSLVKQITKPKRRSTKQCSEIQTPSLPKLIY 320
WD40-rpts
Query 321 SPSRVSESPGSPGQRSQAGEEVTVVFADDAGVHVYASCEDDIYIHTTDSEEEITVIDAASPRPVSLSSDRSVKQSQKLYP 400
WD40-rpts
Query 401 DPILKLNRIIGFGGATMRCAVWSLDGEEVVYPCHAIIVSMTVSSGQQRFFIGHTDKVSALALNASSTVLASAQTGTNSLI 480
WD40-rpts (_________________WD 1________________)(__________________WD 2_______________
Query 481 RLWSYSAGVCQTIIPTHTHALNLLSFSHSGGVLCGVGKDSHNKTTVVLWNTRHVTDGSVVPVLAKAHTDVDIQTIKIAFF 560
WD40-rpts ___) (___________________WD 3__________________)(_____________________WD 4____
Query 561 DDTRMVSCGSGNVRLWRVRGGSLRSCPVNLGQYHNLEFTDLAFEQGHTQNRPVDDRTLFVSSRSGHILEIDYVAVAIKNI 640
WD40-rpts ________________) (________________WD 5________________)
Query 641 RRLLPAHESHTHQREKQTFSTGAGIAVNSISVCAKFCVTGSEDGFMRLWPLDFSSVFLEAEHEGPVSLVNVSSDGSRVLA 720
WD40-rpts (_________________WD 6________________) (_________________WD 7______
Query 721 ATSTGNLGYLDVASRGYRTLMRSHTASVLGFSVDGVRRRVTTVSRDATVRVWDMDTLQQLYDFVTDDDENPCSVAFHPSM 800
WD40-rpts __________) (_________________WD 8________________)(___________________WD 9___
Query 801 PLFACGSSSGAVRVFDIQTSSLIAQHRQHCGSVECVIYSPDGQCLFSAGSGYLVLYSSSEREHHVVRVLGNVVMCGVNRS 880
WD40-rpts _______________) (_________________WD10________________) (___________
Query 881 PDILAVSSDSRRLALIGPTEYIVTIMEARSLNELLRVDVSIVDVESSTLDSALNACFSPASLSHLLISTSANKILWIDTH 960
WD40-rpts ______WD11________________) (_________________WD12________________)
Query 961 TGQLLRQVSEVHKHQCESLAVSEDGHYLLTAGQNSLKVWDYDMKLDVNSQVFIGHSEPIRQVRFTPDQVGVVTAGDALFF 1040
WD40-rpts (_________________WD13________________) (________________WD14______________
Query 1041 WDFLAPPQQSEHCRSPALNVSSRKSAQSWAGLNISELSNEKPREALFHPSSPPVMNFLSTKRNDTEDLLCKSAAELQLSP 1120
WD40-rpts _) (_________________WD15________________) (________________W
Query 1121 SADHAHSPPPRHASFLRVTEWEKDEDTPTLSGHQSGSEKTQGHTLIHPDCYKHFTPRFKTSVVDQSVVASPSGEAGLSLK 1200
WD40-rpts D16________________) (_____________________WD17_____________________)(________
Query 1201 AVIGYNGNGRSNMVWNPDTGLFVYTCGSIVIIEDLHTGSQKQWFGHTEEISTLAVTHDALTVASASGGGASHSSLICIWN 1280
WD40-rpts ___________WD18__________________)(____________________WD19____________________)
Query 1281 VQDGSCRHKLAYHNGAVQSLTFSRDDMYLLSVGDFSEALVALWSTRSFELLCTVQLSIPLHDAAFSPFTANELMLVGSSA 1360
WD40-rpts (__________________WD20_________________)(___________________WD21____________
Query 1361 VVFTRIHTHDNTTELQVQKVGLPDAVGQAEVTSVCYNTRNILYTGTNSGHVCVWDCNTQRCFVTWEADRGEIGVLVCCGN 1440
WD40-rpts _______) (_______________WD22______________) (________________WD23_
Query 1441 MLVTGSNTKKLRLWSVASVQSLSNSDSKRSRAQDDGTQVQLEQELLLDGTIVSAAFDKVMDMGIVGTTAGTLWYINWTDS 1520
WD40-rpts ______________) (_________________WD24________________) (
Query 1521 TSIRLISGHKSKVNGVVCSPDEQHFVTCGQDGSVRVWTLQSHELLVQFQVLNQSCECVCWSPPRGVCGSRGRMCVAAGYS 1600
WD40-rpts _________________WD25________________) (_____________________WD26_____________
Query 1601 DGTVRIFSLDSAEMELKLQPQPDSVCALQFSHYGDVLLSGARDGVITVCSPVTGVSIRTITDHRGASINTLQCTSKQFKE 1680
WD40-rpts _______) (_________________WD27________________) (__________________W
Query 1681 FGLDGNELWLAVSADRRVSIWAADWTDNKCELLDWLTFPAPSPREDLHTLPPSLAAFSPSQRGVVVYTGYAMEKEIIFYS 1760
WD40-rpts D28__________________) (__________________WD29__________________)
Query 1761 LYKKQIVRTISLSDWALSLCLSGKLLAVGSNQRVLNLIHSSSGRFQDFTSHSDSVQVCSFSISGSQLFTAAHNQLLLWTV 1840
WD40-rpts (_________________WD30________________)(_________________WD31_________________)
Query 1841 NGL 1843
WD40-rpts
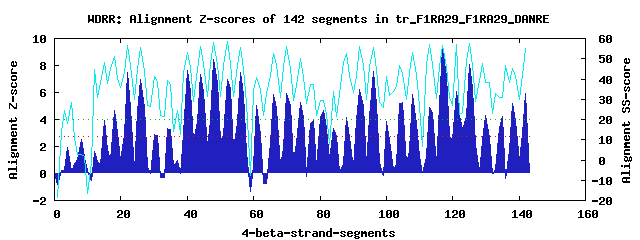 31 WD40-repeats found in QUERY=tr_F1RA29_F1RA29_DANRE (Len=1843): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 18 404-442 4.65 306.91 0.00454 12.5% 50.88 WD 2: Seg 22 443-484 7.51 424.69 0.00012 22.5% 56.52 WD 3: Seg 26 488-530 6.96 402.21 0.00024 30.0% 55.68 WD 4: Seg 30 531-577 2.87 233.25 0.04384 17.5% 41.43 WD 5: Seg 34 585-622 3.33 252.36 0.02446 17.5% 39.19 WD 6: Seg 40 652-690 7.65 430.70 9.8e-05 27.5% 53.05 WD 7: Seg 44 693-731 7.35 418.47 0.00014 22.5% 56.07 WD 8: Seg 48 735-773 8.46 464.10 3.5e-05 27.5% 57.84 WD 9: Seg 52 774-816 6.94 401.42 0.00024 22.5% 58.47 WD10: Seg 56 820-858 7.49 424.14 0.00012 22.5% 55.61 WD11: Seg 61 869-907 5.01 321.47 0.00289 17.5% 40.98 WD12: Seg 66 919-957 5.84 355.96 0.00099 15.0% 52.43 WD13: Seg 70 962-1000 5.94 360.15 0.00087 35.0% 56.25 WD14: Seg 74 1006-1042 5.28 332.75 0.00204 22.5% 50.05 WD15: Seg 78 1061-1099 3.89 275.46 0.01202 10.0% 37.32 WD16: Seg 81 1103-1140 4.68 307.95 0.00440 12.5% 29.20 WD17: Seg 84 1144-1191 3.30 251.08 0.02544 25.0% 33.66 WD18: Seg 88 1192-1234 4.18 287.15 0.00838 22.5% 52.43 WD19: Seg 92 1235-1280 6.25 372.68 0.00059 20.0% 56.11 WD20: Seg 96 1284-1324 7.57 427.54 0.00011 30.0% 55.65 WD21: Seg100 1325-1368 3.83 272.80 0.01305 20.0% 41.40 WD22: Seg105 1381-1415 5.27 332.21 0.00207 25.0% 42.73 WD23: Seg108 1419-1455 5.85 356.15 0.00099 20.0% 53.06 WD24: Seg113 1478-1516 4.89 316.48 0.00338 15.0% 57.00 WD25: Seg117 1520-1558 9.31 499.11 1.2e-05 37.5% 56.44 WD26: Seg121 1562-1608 6.07 365.45 0.00074 22.5% 57.19 WD27: Seg125 1612-1650 8.08 448.39 5.6e-05 32.5% 57.35 WD28: Seg130 1661-1702 4.30 292.08 0.00719 20.0% 38.29 WD29: Seg135 1719-1760 4.22 289.09 0.00789 17.5% 30.74 WD30: Seg138 1761-1799 5.18 328.77 0.00231 15.0% 45.11 WD31: Seg142 1800-1839 5.96 360.88 0.00085 20.0% 54.93Multiple sequence alignment of 31 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 404-442 -L---K---------L-NRIIG-F-GG-ATMRCA---------VWSLD--------GE---EV-V---YPC-----H--AI----IVSMTV WD 2 443-484 SS---G---------Q-QRFFI-G-HT-DKVSAL---------ALNAS--------ST---VL-A---SAQ-----TGTNS----LIRLWS WD 3 488-530 -G---V---------C-QTIIP-T-HT-HALNLL---------SFSHS--------GG---VL-C---GVG-----K--DSHNKTTVVLWN WD 4 531-577 TR---HVTDGSVVPVL-AKAHT-D-VD-IQTIKI---------AFFDD--------TR---MV-S---CGS---------G----NVRLWR WD 5 585-622 -S---C---------P-VNLGQ-Y-HN-LEFTDL---------AFE-Q--------GH---TQ-N---RPV-----D--DR----TLFVSS WD 6 652-690 -H---Q---------R-EKQTF-S-TG-AGIAVN---------SISVC--------AK---FC-V---TGS-----E--DG----FMRLWP WD 7 693-731 -F---S---------S-VFLEA-E-HE-GPVSLV---------NVSSD--------GS---RV-L---AAT-----S--TG----NLGYLD WD 8 735-773 -R---G---------Y-RTLMR-S-HT-ASVLGF---------SVDGV--------RR---RV-T---TVS-----R--DA----TVRVWD WD 9 774-816 MDTLQQ---------L-YDFVT-D-DD-ENPCSV---------AFHPS--------MP---LF-A---CGS-----S--SG----AVRVFD WD10 820-858 -S---S---------L-IAQHR-Q-HC-GSVECV---------IYSPD--------GQ---CL-F---SAG-----S--GY----LVLYSS WD11 869-907 LG---N---------V-V--MC-G-VN-RSPDIL---------AVSSD--------SR---RL-A--LIGP-----T--EY----IVTIME WD12 919-957 -V---S---------I-VDVES-S-TL-DSALNA---------CFSPA--------SL---SH-L---LIS-----T--SA----NKILWI WD13 962-1000 -G---Q---------L-LRQVSEV-HK-HQCESL---------AVSED--------GH---YL-L---TAG--------QN----SLKVWD WD14 1006-1042 -D---V---------N-SQVFI-G-HS-EPIRQV---------RFTPD--------QV---GV-V---TAG-----D--------ALFFWD WD15 1061-1099 -S---S---------R-KSAQS-W-AG-LNISEL---------SNEKP--------RE---AL-F---HPS-----S--PP----VMNFLS WD16 1103-1140 -N---D---------T-EDLLC-K-SA-AEL-QL---------SPSAD--------HA---HS-P---PPR-----H--AS----FLRVTE WD17 1144-1191 -D---E---------D-TPTLS-G-HQ-SGSEKTQGHTLIHPDCYKHF--------TP---RF-K---TSV-----V--DQ----SVVASP WD18 1192-1234 SG---E------AGLS-LKAVI-G-YNGNGRSNM---------VWNPD--------TG---LF-V---YTC--------GS----IVIIED WD19 1235-1280 LH---T--------GS-QKQWF-G-HT-EEISTL---------AVTHD--------AL---TV-A---SASGGGASH--SS----LICIWN WD20 1284-1324 -G---S---------C-RHKLA-Y-HN-GAVQSL---------TFSRD--------DM---YL-L---SVG-----DFSEA----LVALWS WD21 1325-1368 TR---S---------FELLCTV-Q-LS-IPLHDA---------AFSPF--------TANELML-V---GSS-----A--VV----FTRIHT WD22 1381-1415 -----------------GLPDA-V-GQ-AEVTSV---------CYNTR--------N----IL-Y---TGT-----N--SG----HVCVWD WD23 1419-1455 -Q---R---------C-FVTWE-A-DR-GEIGVL---------VCC----------GN---ML-V---TGS-----N--TK----KLRLWS WD24 1478-1516 -Q---V---------Q-LEQEL-L-LD-GTIVSA---------AFDKV--------MD---MG-I---VGT-----T--AG----TLWYIN WD25 1520-1558 -S---T---------S-IRLIS-G-HK-SKVNGV---------VCSPD--------EQ---HF-V---TCG-----Q--DG----SVRVWT WD26 1562-1608 -H---E---------L-LVQFQ-V-LN-QSCECV---------CWSPPRGVCGSRGRM---CV-A---AGY-----S--DG----TVRIFS WD27 1612-1650 -A---E---------M-ELKLQ-P-QP-DSVCAL---------QFSHY--------GD---VL-L---SGA-----R--DG----VITVCS WD28 1661-1702 TD---H---------R-GASIN-TLQC-TSKQFK---------EFGLD--------GN---ELWL---AVS-----A--DR----RVSIWA WD29 1719-1760 -P---A---------P-SPRED-L-HT-LPPSLA---------AFSPS--------QR---GV-VVYTGYA-----M--EK----EIIFYS WD30 1761-1799 LY---K-----------KQIVR-T-IS-LSDWAL---------SLCLS--------GK---LL-A---VGS-----N--QR----VLNLIH WD31 1800-1839 SS---S--------GR-FQDFT-S-HS-DSVQVC---------SFSIS--------GS---QL-F---TAA-----H--N-----QLLLWT WD40 Template TG---E---------C-LRTLS-G-HT-SGVTSV---------SFSPD--------GK---RL-V---SGS-----E--DG----TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |