WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: 89NHE1330741267 Submitted: 2012-03-03 10:21:07 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 408510467 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = 2AQ5_A_PDBID_CHAIN_SEQUENCE (Len=253) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (2AQ5_A_PDBID_CHAIN_SEQUENCE, Len=253): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MSRQVVRSSKFRHVFGQPAKADQCYEDVRVSQTTWDSGFCAVNPKFMALICEASGGGAFLVLPLGKTGRVDKNVPLVCGH 80
WD40-rpts (_________________________WD 1_________________________) (________________
Query 81 TAPVLDIAWCPHNDNVIASGSEDCTVMVWEIPDGGLVLPLREPVITLEGHTKRVGIVAWHPTAQNVLLSAGCDNVILVWD 160
WD40-rpts _____WD 2____________________) (_________________WD 3_________________)
Query 161 VGTGAAVLTLGPDVHPDTIYSVDWSRDGALICTSCRDKRVRVIEPRKGTVVAEKDRPHEGTRPVHAVFVSEGKILTTGFS 240
WD40-rpts (__________________WD 4_________________) (___________________WD 5_________
Query 241 RMSERQVALWDTK 253
WD40-rpts __________)
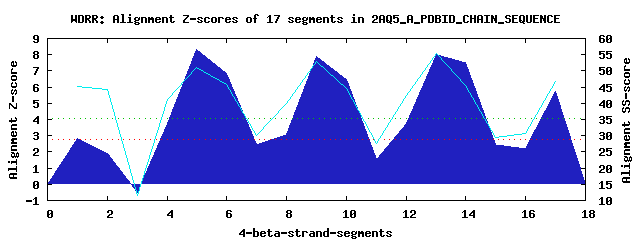 5 WD40-repeats found in QUERY=2AQ5_A_PDBID_CHAIN_SEQUENCE (Len=253): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 7-62 2.84 231.83 0.04577 15.0% 45.34 WD 2: Seg 5 64-110 8.30 457.68 4.2e-05 35.0% 51.11 WD 3: Seg 9 121-160 7.87 439.87 7.3e-05 40.0% 52.75 WD 4: Seg 13 164-204 8.02 445.96 6.1e-05 27.5% 55.25 WD 5: Seg 17 208-251 5.74 351.93 0.00112 25.0% 46.62Multiple sequence alignment of 5 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 7-62 RSS------KFRHVFGQPAKADQCYEDVRVS--QTTWDSGFCA--VNP-KFMA-LICE---ASGGGAFLVL WD 2 64-110 LGKTGRVDKNVP----------------LVC--GHTAPVLDIA--WCPHNDNV-IASG---SEDCTVMVWE WD 3 121-160 -RE------PVI----------------TLE--GHTKRVGIVA--WHP-TAQNVLLSA---GCDNVILVWD WD 4 164-204 -GA------AVL----------------TLGPDVHPDTIYSVD--WSR-DGAL-ICTS---CRDKRVRVIE WD 5 208-251 -GT------VVA----------------EKD--RPHEGTRPVHAVFVS-EGKI-LTTGFSRMSERQVALWD WD40 Template TGE------CLR----------------TLS--GHTSGVTSVS--FSP-DGKR-LVSG---SEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |