WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: 88EDP1659460623 Submitted: 2022-08-03 01:17:03 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 79790783 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CmaCh03G010850 (Len=661) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CmaCh03G010850, Len=661): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MLSSDGDADVFFDSEDYLPLEESLVTEEVDVSKSGYEIWMREPQSIKKRREYFLQEVGLKEFASSSSTRTQDSEIESSCS 80
WD40-rpts (_________________WD 1_________________)(_________________WD 2________________)
Query 81 SGRMELERLTECSGAVSSSDYAPSCNVVDNDSVVHHNDMQSDEFSNEANEATSRPSSVSNSSFQRCQGEANVEACKDIDA 160
WD40-rpts (_______________WD 3_______________) (_________________WD 4________________)
Query 161 RKKKILTWWKSLKNKAARLRAMEFSEESKSNPAKSESCRMKVCQNKKKFSEFSALYMRQQICAHKGLIWTMKFSPDGKYL 240
WD40-rpts (___________________WD 5__________________) (_________________WD 6____
Query 241 ASGGEDGVVRIWRVTRTNASSEFLANDATREGKSSFSSKPLRFATVVIPAKVFQIDELPIQEFHGHSSDVLDIAWSTSNC 320
WD40-rpts ____________) (_________________WD 7________________)(________________WD 8___
Query 321 LLSSSKDKTVRLWQVGSDQCLNVFHHKNYVTCIQFNPMDENYFVSGSIDGKIRIWGVLKQRVVDWADIRDAITAISYQPD 400
WD40-rpts _____________)(__________________WD 9__________________) (_________________WD10
Query 401 GKGFAVGSITGTCRFYKTSGDDYINLDEQINIGGRNKASGKRITGIQFFRENSHRVMITSEDSKVRIFEGTKIVGQYKGL 480
WD40-rpts ________________) (_________________WD11________________) (_______
Query 481 SKSGSQMSASFTENGKHIVSVGEDSRVYMWNYDSVGLSSTNETKSQQSCEHFFSDGVSVAIPCPGTGTEPKCLGNSQSQL 560
WD40-rpts __________WD12________________) (__________________WD13__________________)
Query 561 SQRRDNQGDASGLECFSFSNWFSSNGLRRGSATWPEERLPLWDLPVAEDEHQDCHQKKLNGHNGDAHDQGSRLETWGLVI 640
WD40-rpts (_________________WD14_________________) (__________________WD15_____
Query 641 VVAGSDGTIKTFHNYGLPIKF 661
WD40-rpts ____________)
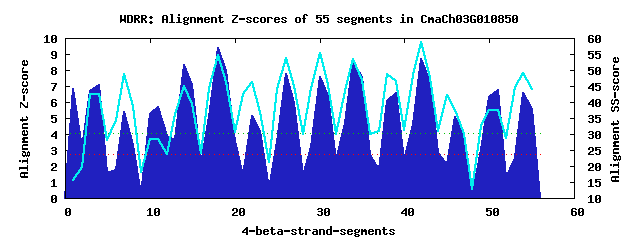 15 WD40-repeats found in QUERY=CmaCh03G010850 (Len=661): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-40 6.80 395.40 0.00029 2.5% 15.71 WD 2: Seg 4 41-79 7.06 406.44 0.00021 10.0% 42.39 WD 3: Seg 7 83-118 5.37 336.37 0.00182 17.5% 48.64 WD 4: Seg 11 122-160 5.66 348.32 0.00126 17.5% 28.48 WD 5: Seg 14 162-204 8.29 457.29 4.3e-05 22.5% 44.88 WD 6: Seg 18 215-253 9.36 501.42 1.1e-05 35.0% 54.63 WD 7: Seg 22 258-296 5.13 326.56 0.00247 15.0% 46.34 WD 8: Seg 26 297-334 7.73 434.15 8.8e-05 27.5% 53.77 WD 9: Seg 30 335-376 7.58 427.66 0.00011 37.5% 55.29 WD10: Seg 34 379-417 8.53 467.05 3.2e-05 25.0% 53.48 WD11: Seg 39 431-469 6.54 384.97 0.00040 15.0% 46.54 WD12: Seg 42 473-511 8.69 473.56 2.6e-05 25.0% 58.78 WD13: Seg 46 514-555 5.04 323.00 0.00276 15.0% 37.36 WD14: Seg 51 564-603 6.77 394.49 0.00030 25.0% 37.58 WD15: Seg 54 613-653 6.58 386.49 0.00038 25.0% 48.95Multiple sequence alignment of 15 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-40 ML-SSD---GDADVFFDSE--DYLP-LEE-SLVTEE---VDV-SKSGYEIWM WD 2 41-79 -R-EPQ---SIKKRREYFL--QEVG-LKE-FASSSS---TRT-QDSEIESSC WD 3 83-118 -R-MEL---ERLTECSGAV--SSSD-YAP-SCNVVD---N----DSVVHHND WD 4 122-160 -D-EFS---NEANEATSRP--SSVS-NSS-FQRCQG---EAN-VEACKDIDA WD 5 162-204 KK-KILTWWKSLKNKAARL--RAME-FSE-ESKSNP---AKS-ESCRMKVCQ WD 6 215-253 -L-YMR---QQICAHKGLI--WTMK-FSP-DGKYLA---SGG-EDGVVRIWR WD 7 258-296 -N-ASS---EFLANDATRE--GKSS-FSS-KPLRFA---TVV-IPAKVFQID WD 8 297-334 -E-LPI---QEFHGHSSDV--LDIA-WST-SN-CLL---SSS-KDKTVRLWQ WD 9 335-376 VGSDQC---LNVFHHKNYV--TCIQ-FNPMDENYFV---SGS-IDGKIRIWG WD10 379-417 -K-QRV---VDWADIRDAI--TAIS-YQP-DGKGFA---VGS-ITGTCRFYK WD11 431-469 ---NIG---GRNKASGKRI--TGIQFFRE-NSHRVM---ITS-EDSKVRIFE WD12 473-511 -I-VGQ---YKGLSKSGSQ--MSAS-FTE-NGKHIV---SVG-EDSRVYMWN WD13 514-555 -S-VGL---SSTNETKSQQ--SCEH-FFS-DGVSVAIPCPGT-GTEPKCLGN WD14 564-603 R--DNQ---GDASGLECFS--FSNW-FSS-NGLRRG---SATWPEERLPLWD WD15 613-653 -D-CHQ---KKLNGHNGDAHDQGSR-LET-WGLVIV---VAG-SDGTIKTFH WD40 Template TG-ECL---RTLSGHTSGV--TSVS-FSP-DGKRLV---SGS-EDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |