WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| JobID: 0ZDQY1349454144 Submitted: 2012-10-06 00:22:24 You can bookmark this page for further accessing the result in the future. Results on this server will be retained for 30 days. Job status: [Done] Total run time: 389797234 seconds. [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = gi_197927448_ref_NP_060501.3_WDR6 (Len=1151) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (gi_197927448_ref_NP_060501.3_WDR6, Len=1151): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MGSAARWEGSFSREFSSLLSRGCLSTSRIDMDALEDYVWPRATSELILLPVTGLECVGDRLLAGEGPDVLVYSLDFGGHL 80
WD40-rpts (_____________WD 1____________)
Query 81 RMIKRVQNLLGHYLIHGFRVRPEPNGDLDLEAMVAVFGSKGLRVVKISWGQGHFWELWRSGLWNMSDWIWDARWLEGNIA 160
WD40-rpts (_________________WD 2_________________)(___________________WD 3________
Query 161 LALGHNSVVLYDPVVGCILQEVPCTDRCTLSSACLIGDAWKELTIVAGAVSNQLLVWYPATALADNKPVAPDRRISGHVG 240
WD40-rpts ___________) (___________________WD 4__________________) (___________
Query 241 IIFSMSYLESKGLLATASEDRSVRIWKVGDLRVPGGRVQNIGHCFGHSARVWQVKLLENYLISAGEDCVCLVWSHEGEIL 320
WD40-rpts ______WD 5________________)(_____________________WD 6____________________) (___
Query 321 QAFRGHQGRGIRAIAAHERQAWVITGGDDSGIRLWHLVGRGYRGLGVSALCFKSRSRPGTLKAVTLAGSWRLLAVTDTGA 400
WD40-rpts ______________WD 7_________________) (________________WD 8____________
Query 401 LYLYDVEVKCWEQLLEDKHFQSYCLLEAAPGPEGFGLCAMANGEGRVKVVPINTPTAAVDQTLFPGKVHSLSWALRGYEE 480
WD40-rpts ____) (_________________WD 9_________________) (__________________WD10___
Query 481 LLLLASGPGGVVACLEISAAPSGKAIFVKERCRYLLPPSKQRWHTCSAFLPPGDFLVCGDRRGSVLLFPSRPGLLKDPGV 560
WD40-rpts _______________) (_______________________WD11______________________)(__________
Query 561 GGKARAGAGAPVVGSGSSGGGNAFTGLGPVSTLPSLHGKQGVTSVTCHGGYVYTTGRDGAYYQLFVRDGQLQPVLRQKSC 640
WD40-rpts _______WD12_________________)(_______________WD13_______________) (___________
Query 641 RGMNWLAGLRIVPDGSMVILGFHANEFVVWNPRSHEKLHIVNCGGGHRSWAFSDTEAAMAFAYLKDGDVMLYRALGGCTR 720
WD40-rpts ________WD14__________________) (___________
Query 721 PHVILREGLHGREITCVKRVGTITLGPEYGVPSFMQPDDLEPGSEGPDLTDIVITCSEDTTVCVLALPTTTGSAHALTAV 800
WD40-rpts ______WD15________________)(_________________WD16________________) (______
Query 801 CNHISSVRAVAVWGIGTPGGPQDPQPGLTAHVVSAGGRAEMHCFSIMVTPDPSTPSRLACHVMHLSSHRLDEYWDRQRNR 880
WD40-rpts ___________WD17________________)
Query 881 HRMVKVDPETRYMSLAVCELDQPGLGPLVAAACSDGAVRLFLLQDSGRILQLLAETFHHKRCVLKVHSFTHEAPNQRRRL 960
WD40-rpts (_______________WD18_______________) (_______________________WD19________
Query 961 LLCSAATDGSLAFWDLTTMLDHDSTVLEPPVDPGLPYRLGTPSLTLQAHSCGINSLHTLPTREGHHLVASGSEDGSLHVF 1040
WD40-rpts ______________) (__________________WD20__________________
Query 1041 VLAVEMLQLEEAVGEAGLVPQLRVLEEYSVPCAHAAHVTGLKILSPSIMVSASIDQRLTFWRLGHGEPTFMNSTVFHVPD 1120
WD40-rpts ) (_______________WD21_______________) (_______
Query 1121 VADMDCWPVSPEFGHRCALGGQGLEVYNWYD 1151
WD40-rpts __________WD22________________)
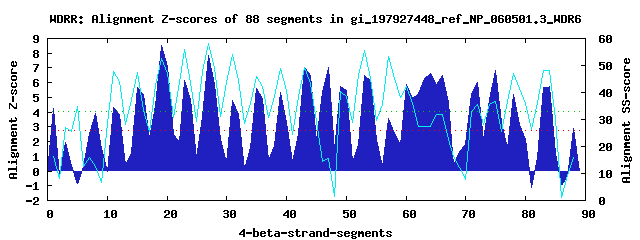 22 WD40-repeats found in QUERY=gi_197927448_ref_NP_060501.3_WDR6 (Len=1151): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-31 4.22 288.94 0.00792 17.5% 16.15 WD 2: Seg 8 89-128 3.88 275.07 0.01216 7.5% 12.48 WD 3: Seg 11 129-172 4.32 293.22 0.00694 7.5% 47.82 WD 4: Seg 15 176-218 5.68 349.48 0.00121 17.5% 47.46 WD 5: Seg 19 229-267 8.54 467.30 3.1e-05 27.5% 52.59 WD 6: Seg 23 268-314 6.14 368.39 0.00067 27.5% 55.86 WD 7: Seg 27 317-356 7.85 438.81 7.6e-05 25.0% 58.25 WD 8: Seg 31 368-405 4.77 311.74 0.00391 12.5% 54.00 WD 9: Seg 35 412-451 5.58 344.99 0.00139 17.5% 46.01 WD10: Seg 39 455-496 5.33 334.68 0.00192 15.0% 48.91 WD11: Seg 43 499-549 7.04 405.31 0.00021 20.0% 49.42 WD12: Seg 47 550-589 7.06 406.37 0.00021 12.5% 15.61 WD13: Seg 50 590-625 5.50 341.73 0.00154 15.0% 38.69 WD14: Seg 53 629-671 6.51 383.55 0.00042 20.0% 55.58 WD15: Seg 60 709-747 5.88 357.44 0.00095 20.0% 42.04 WD16: Seg 64 748-786 6.59 386.77 0.00038 12.5% 27.48 WD17: Seg 66 794-832 6.49 382.85 0.00043 15.0% 31.72 WD18: Seg 72 887-922 6.03 363.89 0.00078 12.5% 35.65 WD19: Seg 75 925-975 6.85 397.55 0.00027 22.5% 36.79 WD20: Seg 78 1000-1041 5.18 328.58 0.00232 35.0% 47.03 WD21: Seg 84 1067-1102 5.74 351.90 0.00113 20.0% 48.17 WD22: Seg 88 1113-1151 2.92 235.44 0.04102 10.0% 16.09Multiple sequence alignment of 22 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-31 MG-----------------------------------S-----------A-ARWE----GSFSREFS---SL-LSRGCLSTSRIDM WD 2 89-128 LL---------G------HYLIH-----------GFRV-----------R-PEPN----GDLDLEAM---VA-VFGSKGLRVVKIS WD 3 129-172 WG---------QGHFWELWRSGL-----------WNMS-----------D-WIWD----ARWLEGN-----I-ALALGHNSVVLYD WD 4 176-218 -G---------C------ILQEV-----------PCTD-----------R-CTLSSACLIGDAWKEL---TI-VAGAVSNQLLVWY WD 5 229-267 -V---------A------PDRRI-----------SGHV-----------G-IIFS----MSYLESKG---LL-ATASEDRSVRIWK WD 6 268-314 VGDLRVPGGRVQ------NIGHC-----------FGHS-----------A-RVWQ----VKLLEN-----YL-ISAGEDCVCLVWS WD 7 317-356 -G---------E------ILQAF-----------RGHQ-----------GRGIRA----IAAHERQA---WV-ITGGDDSGIRLWH WD 8 368-405 -S---------A------LCFKS-----------RSRP-----------G-TLKA----VTLAGSW----RL-LAVTDTGALYLYD WD 9 412-451 -E---------Q------LLEDK-----------HFQS-----------Y-CLLE----AAPGPEGF---GLCAMANGEGRVKVVP WD10 455-496 -P---------T------AAVDQ-----------TLFP-----------G-KVHS----LSWALRGYEELLL-LASGPGGVVACLE WD11 499-549 AA---------P------SGKAIFVKERCRYLLPPSKQ-----------R-WHTC----SAFLPPGD---FL-VCGDRRGSVLLFP WD12 550-589 SR---------P------GLLKD-----------PGVG-----------G-KARA----GAGAPVVG---SG-SSGGGNAFTGLGP WD13 590-625 -------------------VSTL-----------PSLH-----------G-KQGV----TSVTCHGG---YV-YTTGRDGAYYQLF WD14 629-671 -G---------Q------LQPVL-----------RQKS-------CRGMN-WLAG----LRIVPDGS---MV-ILGFHANEFVVWN WD15 709-747 -V---------M------LYRAL-----------GGCT-----------R-PHVI----LREGLHGR---EI-TCVKRVGTITLGP WD16 748-786 -E---------Y------GVPSF-----------MQPD-----------D-LEPG----SEGPDLTD---IV-ITCSEDTTVCVLA WD17 794-832 -A---------H------ALTAV-----------CNHI-----------S-SVRA----VAVWGIGT---PG-GPQDPQPGLTAHV WD18 887-922 DP---------E----------T-----------RYMS-----------L-AVCE----LDQPGLGP---LV-AAACSDGAVRLFL WD19 925-975 DS---------G------RILQL-----------LAETFHHKRCVLKVHS-FTHE----APNQRRRL---LL-CSAATDGSLAFWD WD20 1000-1041 -G---------T------PSLTL-----------QAHS-----------C-GINS----LHTLPTREGHHLV-ASGSEDGSLHVFV WD21 1067-1102 ------------------EYSVP-----------CAHA-----------A-HVTG----LKIL-SPS---IM-VSASIDQRLTFWR WD22 1113-1151 ST----------------VFHVP-----------DVAD-----------M-DCWP----VSPEFGHR---CA-LGGQGLEVYNWYD WD40 Template TG---------E------CLRTL-----------SGHT-----------S-GVTS----VSFSPDGK---RL-VSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |