WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: YPR169W [Saccharomyces cerevisiae] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = YPR169W (Len=492) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (YPR169W, Len=492): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAKSKKKTDVVDSTNLPILELLSLKAPIFQSLLHPELPIIITGFGTGHIVCHRYDPAKLQSHLDRRRRIDTATTGKDAKK 80
WD40-rpts (_________________WD 1________________)
Query 81 GVCPWIRLDIDLETGDLKFVDIEEQQQQKQTGKDEDLGVKTLWKTKRHKGSVRAMCFDSKGDNIFSVGSDNVLKKANTMT 160
WD40-rpts (_______________________WD 2______________________)
Query 161 GKVVKKVNLSSLFNSEEKKNDKFTKLCASQTHPFILIGDESGNIHVINSENLALSNSIRSIHFGDSINDIFHFDKRSAYK 240
WD40-rpts (_________________WD 3_________________) (__________________WD 4______
Query 241 FISLGQTTLAYFDVRDKDAKPNVAGNEDGKILISDDQEDEVLCGCFVDPEVADTLLCGMGEGIVTVWKPNKNDLEDQMSR 320
WD40-rpts ____________) (_______________________WD 5_______________________) (____
Query 321 IKISKDESIDCIVPTLQDDNCVWCGCSNGNIYKVNAKLGKVVEIRNHSELDEVSFVDLDFEYRVVSGGLENIKIWELSSD 400
WD40-rpts _____________WD 6_________________)(__________________WD 7_________________) (
Query 401 DVEENASVESDSDEPLSHSDEDLSDDTSSDDETTLVGLSKEELLDELDKDLKEDHQEEKESNSKSVKKRKIMKENNKKKD 480
WD40-rpts _________________WD 8________________)
Query 481 LYEHGIKKFDDL 492
WD40-rpts
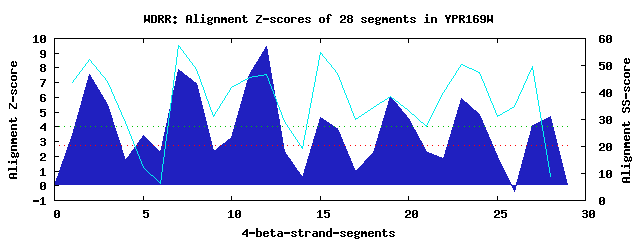 8 WD40-repeats found in QUERY=YPR169W (Len=492): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 15-53 7.55 426.58 0.00011 12.5% 52.36 WD 2: Seg 7 107-157 7.87 439.76 7.4e-05 20.0% 57.29 WD 3: Seg 12 169-208 9.45 505.20 9.7e-06 17.5% 46.49 WD 4: Seg 15 212-253 4.64 306.13 0.00465 7.5% 54.67 WD 5: Seg 19 257-308 6.03 363.95 0.00077 20.0% 38.52 WD 6: Seg 23 316-355 5.92 359.19 0.00090 15.0% 50.37 WD 7: Seg 27 356-396 4.08 283.26 0.00945 15.0% 49.71 WD 8: Seg 28 400-438 4.73 310.23 0.00410 17.5% 8.87Multiple sequence alignment of 8 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 15-53 -NL-----------PILELLS--LKAPIFQ-SL-------------LHPELPIIITGFGTGHIVCHR WD 2 107-157 QQKQTGKDEDLGVKTLWKTKR--HKGSVRA-MC-------------FDSKGDNIFSVGSDNVLKKAN WD 3 169-208 LSS-----------LFNSEEK--KNDKFTK-LC-------------ASQTHPFILIGDESGNIHVIN WD 4 212-253 -LA-----------LSNSIRSIHFGDSIND-IF------------HFDKRSAYKFISLGQTTLAYFD WD 5 257-308 -KD-----------AKPNVAG--NEDGKIL-ISDDQEDEVLCGCFVDPEVADTLLCGMGEGIVTVWK WD 6 316-355 -DQ-----------MSRIKIS--KDESIDCIVP-------------TLQDDNCVWCGCSNGNIYKVN WD 7 356-396 AKL----------GKVVEIRN--HSELDEV-SF-------------VDLDFEYRVVSGGLENIKIWE WD 8 400-438 -DD-----------VEENASV--ESDSDEP-LS-------------HSDEDLSDDTSSDDETTLVGL WD40 Template TGE-----------CLRTLSG--HTSGVTS-VS-------------FSPDGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |