WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: YPL183C [Saccharomyces cerevisiae] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = YPL183C (Len=1013) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (YPL183C, Len=1013): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MKDLSHYGPALCVKFYNDYVLAGYGPFIHVYDYHSATLINKCRLFHYNKVHGLSLSSEGKILAYGARSVTIVELEDVLKK 80
WD40-rpts (_____________WD 1_____________)(___________________WD 2___________________)
Query 81 ESLVDFERINSDWITGATFSFDNLQIYLLTCYNKVLICDLNCEVLFRKSLGGERSILYSGIIKVFGPDKVYVNAGTVMGG 160
WD40-rpts (_________________WD 3________________) (_______________WD 4___________
Query 161 VIIWDLFSETKIHNLLGHEGSIFYVNLSNNGRYVASCSDDRSIRLWDLETGKQLSVGWSHTARIWNLMFFDNDSKLISVS 240
WD40-rpts ____) (_________________WD 5________________) (_________________WD 6________
Query 241 EDCTCRVWNIIESRENVAELSISNVYEVHLIKSIWGVDVKDDEMIAVTSGNDGRLKLIDLLQLKRHGDEETSFSLDDIAK 320
WD40-rpts ________) (_________________WD 7_________________)(________________WD 8
Query 321 QCGDIFEKNESIKGFQWFSFGVIAITSLGKILKYSDVTKQWKLLLTNEKFNSYPITNGIQTQNIAVFSNNKSDILLIKFS 400
WD40-rpts _______________) (________________WD 9________________) (_________________WD
Query 401 KDSADIIETEEFHLDELSKTNNCLVTEYDDDSFLLTLQSPNPREKFVCLEISLQNLKIKSKHCFNKPENFSSSCLTSFRN 480
WD40-rpts 10________________) (__________________WD11___
Query 481 HILVGSRFSTLVIYNLLDESEEPFIIRRLSPGDTTTSIEFVEDKDNSAVFSVTNRDGYYVFIELTKNSLEEGPYRLSYKV 560
WD40-rpts ______________) (_______________WD12_______________) (________
Query 561 LHSNKMMKGFLEGAFFNSKGEYITYGFKSSLFYLYNETNCYELASEVCGGSHRLWNLAKITDGHVLMYIKASRFHLRKIY 640
WD40-rpts ____________WD13___________________)(__________________WD14__________________)
Query 641 NSIVPETLENGVHGREIRDISICPVSNTNTNDNFKDGHIFCTASEDTTIKLGYFNNRTGKVQNFWTQRKHVSGLQRCQFI 720
WD40-rpts (____________________WD15___________________) (________________WD1
Query 721 NHKLMISSSAREELFLWELNDKYNKRPYMTIRQALPVSTNNSDLRIMDFDVKFISQSGDFLLVTVYSDSTIKIWHYRENQ 800
WD40-rpts 6________________) (_________________WD17________________) (__
Query 801 NKFDLIMQGRYKTCCLFNVVFIALKEELLVVISPTDGHLVVYNITEYVPFSVDPISGDLVDHKLDATISNLPAPVAQLPV 880
WD40-rpts __________________WD18____________________) (________
Query 881 HQSGVKSLDYVANATRTSATILTGGDDNGLGLSNLKLDDSNKVTLKTSDFIAAAASSTITSGMLINGGKEVITTSVDQVI 960
WD40-rpts ___________WD19__________________) (_____________________WD20__________________
Query 961 RAWEITAGKLSLVDKKRTTVADTGSLEIISNDEDADSEKTLLIGGVGLSIWKK 1013
WD40-rpts ___)
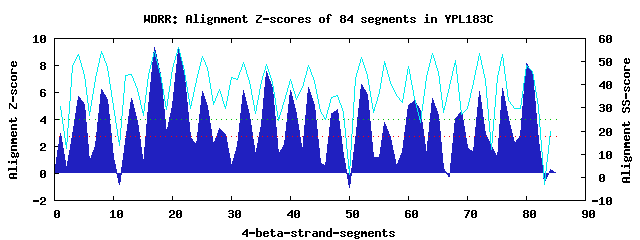 20 WD40-repeats found in QUERY=YPL183C (Len=1013): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-32 2.93 235.55 0.04088 20.0% 30.71 WD 2: Seg 4 33-76 5.69 349.82 0.00120 27.5% 53.03 WD 3: Seg 8 81-119 6.20 370.58 0.00063 12.5% 54.23 WD 4: Seg 13 130-165 5.53 342.90 0.00149 27.5% 44.37 WD 5: Seg 17 169-207 9.34 500.28 1.1e-05 30.0% 54.24 WD 6: Seg 21 211-249 9.22 495.53 1.3e-05 32.5% 55.94 WD 7: Seg 25 260-299 6.07 365.50 0.00074 20.0% 52.25 WD 8: Seg 29 300-336 2.88 233.57 0.04342 27.5% 29.58 WD 9: Seg 32 340-377 6.12 367.66 0.00069 7.5% 49.79 WD10: Seg 36 381-419 7.54 426.29 0.00011 15.0% 48.79 WD11: Seg 43 455-495 6.39 378.51 0.00049 10.0% 48.30 WD12: Seg 48 508-543 4.72 309.48 0.00420 10.0% 35.36 WD13: Seg 52 552-596 6.60 387.29 0.00038 10.0% 51.69 WD14: Seg 56 597-638 3.81 272.20 0.01329 12.5% 50.21 WD15: Seg 61 649-693 5.40 337.65 0.00175 25.0% 33.87 WD16: Seg 64 701-738 5.57 344.83 0.00140 22.5% 53.39 WD17: Seg 69 757-795 4.56 302.82 0.00516 25.0% 27.11 WD18: Seg 72 798-843 6.07 365.60 0.00074 17.5% 53.35 WD19: Seg 76 872-914 6.30 375.04 0.00055 20.0% 53.21 WD20: Seg 80 917-964 8.16 451.94 5.0e-05 22.5% 48.26Multiple sequence alignment of 20 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-32 ----------------MKD--------LSH-YGPALC---VKF----YNDYVLAGYGP------FI-HVYD WD 2 33-76 YH-----SATLI---NKCR--------LFH-YNKVHG---LSL----SSEGKILAYGA---RSVTIVELED WD 3 81-119 -E-----S---L---VDFE--------RIN-SDWITG---ATF----SFDNLQIYLLT---CYNKV-LICD WD 4 130-165 ------------------L--------GGE-RSILYSGIIKVF----GPDKVYVNAGT---VMGGV-IIWD WD 5 169-207 -E-----T---K---IHNL--------LGH-EGSIFY---VNL----SNNGRYVASCS---DDRSI-RLWD WD 6 211-249 -G-----K---Q---LSVG--------WSH-TARIWN---LMF----FDNDSKLISVS---EDCTC-RVWN WD 7 260-299 -L-----S---I---SNVY--------EVHLIKSIWG---VDV----KDDEMIAVTSG---NDGRL-KLID WD 8 300-336 ---------------LLQL--------KRH---GDEE---TSF----SLDDIAKQCGDIFEKNESI-KGFQ WD 9 340-377 FG-------------VIAI--------TSL-GKILKY---SDV----TKQWKLLLTNE---KFNSY-PITN WD10 381-419 -T-----Q---N---IAVF--------SNN-KSDILL---IKF----SKDSADIIETE---EFHLD-ELSK WD11 455-495 NL-----K---I--KSKHC--------FNK-PENFSS---SCL----TSFRNHILVGS---RFSTL-VIYN WD12 508-543 ---------------RLSP--------GDT-TTSIEF---VED----KDNSAVFSVTN---RDGYY-VFIE WD13 552-596 GPYRLSYK---V---LHSN--------KMM-KGFLEG---AFF----NSKGEYITYGF---KSSLF-YLYN WD14 597-638 ET-----N---CYELASEV--------CGG-SHRLWN---LAK----ITDGHVLMYIK----ASRF-HLRK WD15 649-693 ENGVHGRE---I---RDIS--------ICP-VSNTNT---NDN----FKDGHIFCTAS---EDTTI-KLGY WD16 701-738 -V-----Q---N---FWTQ--------RKH-VSGLQR---CQF----INH-KLMISSS---AREEL-FLWE WD17 757-795 VS-----T---N---NSDL--------RIM-DFDVKF---IS-----QSGDFLLVTVY---SDSTI-KIWH WD18 798-843 EN-QNKFD---L---IMQG--------RYK-TCCLFN---VVF--IALKEELLVVISP---TDGHL-VVYN WD19 872-914 -P-----A---P---VAQL--------PVH-QSGVKS---LDYVANATRTSATILTGG---DDNGL-GLSN WD20 917-964 LD-----D---S---NKVTLKTSDFIAAAA-SSTITS---GML----INGGKEVITTS---VDQVI-RAWE WD40 Template TG-----E---C---LRTL--------SGH-TSGVTS---VSF----SPDGKRLVSGS---EDGTI-KVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |