WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: YOR269W [Saccharomyces cerevisiae] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = YOR269W (Len=494) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (YOR269W, Len=494): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MTNWQQQLPLTDTQKNELDKSVLRYLNWNYKQTVRHEHAQDYESVRHAIVTLSGFLLQESVDRQEFISNNDTSNESMVDI 80
WD40-rpts
Query 81 DELLLPKKWNSIVRLQKKIIELEQNTETLVSQIKDLNTQVSELAQFKPTTSNGTSAHNVLKWIPRNLPSCLINVESSVTS 160
WD40-rpts (________________________WD 1________________________) (_______________
Query 161 VKLHPNLPIVFVATDHGKLYAFDLFNYTIPLASLQSHTKAITSMDVLFTNYTNSSKKNYLVIVTASKDLQIHVFKWVSEE 240
WD40-rpts __WD 2________________)(_______________________WD 3_______________________)
Query 241 CKFQQIRSLLGHEHIVSAVKIWQKNNDVHIASCSRDQTVKIWDFHNGWSLKTFQPHSQWVRSIDVLGDYIISGSHDTTLR 320
WD40-rpts (__________________WD 4_________________) (________________WD 5_____________
Query 321 LTHWPSGNGLSVGTGHEFPIEKVKFIHFIEDSPEIRFRTPSTDRYKNWGMQYCVSASRDRTIKIWEIPLPTLMAHRAPIP 400
WD40-rpts __) (___________________________WD 6___________________________)
Query 401 NPTDSNFRCVLTLKGHLSWVRDISIRGQYLFSCADDKSVRCWDLNTGQCLHVWEKLHTGFVNCLDLDVDFDSNVTPRQMM 480
WD40-rpts (________________WD 7_______________) (_____________________WD 8________
Query 481 VTGGLDCKSNVFMR 494
WD40-rpts ____________)
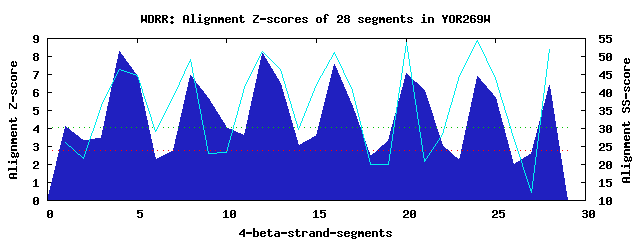 8 WD40-repeats found in QUERY=YOR269W (Len=494): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 90-143 4.09 283.57 0.00936 12.5% 26.17 WD 2: Seg 4 145-183 8.30 457.44 4.3e-05 22.5% 46.51 WD 3: Seg 8 184-235 6.94 401.19 0.00024 30.0% 49.27 WD 4: Seg 12 243-283 8.18 452.64 4.9e-05 32.5% 51.37 WD 5: Seg 16 287-323 7.53 425.66 0.00011 30.0% 50.98 WD 6: Seg 20 327-386 7.07 406.51 0.00021 35.0% 54.01 WD 7: Seg 24 407-443 6.88 398.99 0.00026 35.0% 54.40 WD 8: Seg 28 447-493 6.37 377.75 0.00050 27.5% 51.86Multiple sequence alignment of 8 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 90-143 -NS---IVRLQKKIIELEQNTETLVSQIKD-LNTQVSEL---------AQ--FK---------------------P-------T--TSNGTSAHNVLKWI WD 2 145-183 -RN---LPS-----------C------LIN-VESSVTSV---------KL--HP---------------------N-------LPIVFVATDHGKLYAFD WD 3 184-235 LFNYTIPLA-----------S------LQS-HTKAITSMDVLFTNYTNSS--KK---------------------N-------YLVIVTASKDLQIHVFK WD 4 243-283 -FQ---QIR-----------S------LLG-HEHIVSAV---------KIWQKN---------------------N-------DVHIASCSRDQTVKIWD WD 5 287-323 -GW---SLK-----------T------FQP-HSQWVRSI---------DV--L------------------------------GDYIISGSHDTTLRLTH WD 6 327-386 -GN---GLS-----------V------GTG-HEFPIEKV---------KF--IHFIEDSPEIRFRTPSTDRYKNWG-------MQYCVSASRDRTIKIWE WD 7 407-443 -FR---CVL-----------T------LKG-HLSWVRDI---------SI-------------------------R-------GQYLFSCADDKSVRCWD WD 8 447-493 -GQ---CLH-----------V------WEKLHTGFVNCL---------DL--DV---------------------DFDSNVTPRQMMVTGGLDCKSNVFM WD40 Template TGE---CLR-----------T------LSG-HTSGVTSV---------SF--SP---------------------D-------GKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |