WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: YGR223C [Saccharomyces cerevisiae] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = YGR223C (Len=448) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (YGR223C, Len=448): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MDVRRPIREAVNNRRKPKFLSVSFNQDDSCFSVALENGFRIFNTDPLTSKLSKTFKESATNQSRGTGIGYTRMLYRTNYI 80
WD40-rpts (_______________WD 1_______________)
Query 81 ALVGGGKRPRHALNKLIIWDDLLQKETITLKFMSSIKDVFLSRIHIVVVLENTIEIFQFQTNPQRICPILDIPPNGSVDY 160
WD40-rpts (________________WD 2________________)
Query 161 VVCSSKHLQSQASQSQSKILEIIAFPSNKCVGQIQVADLSQIKYNSQNPKESALLPTSIIKAHKNPIKLVRLNRQGTMVA 240
WD40-rpts (_________________________WD 3____________
Query 241 TCSVQGTLIRIFSTHNGTLIKEFRRGVDKADIYEMSFSPNGSKLAVLSNKQTLHIFQIFETTNTETNTPDHSRANGSSHP 320
WD40-rpts ____________) (__________________WD 4_________________)(___________________WD
Query 321 LKNYIPKGLWRPKYLDSVWSICNAHLKNPIFDAHRNDNSGDVTHDNEFYKDRCRIGWCQDSNNREQDDSLVLVWQNSGIW 400
WD40-rpts 5__________________) (____________________WD 6____________________)
Query 401 EKFVILEKEQQDSSKTHYSLNESLRNEDTKSAGEPTRWELVRESWREL 448
WD40-rpts
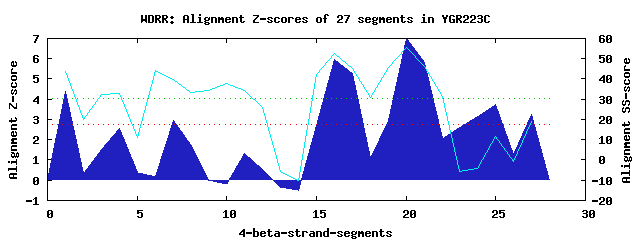 6 WD40-repeats found in QUERY=YGR223C (Len=448): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 8-43 4.37 295.22 0.00652 17.5% 43.84 WD 2: Seg 7 101-138 2.93 235.78 0.04060 17.5% 39.66 WD 3: Seg 16 199-253 5.95 360.63 0.00086 17.5% 52.52 WD 4: Seg 20 257-297 6.98 402.95 0.00023 22.5% 55.74 WD 5: Seg 24 298-340 3.16 245.17 0.03049 15.0% -4.19 WD 6: Seg 25 350-395 3.72 268.25 0.01501 12.5% 11.42Multiple sequence alignment of 6 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 8-43 -------------------REAV---NN-RRK--PKFLSVSFNQDDS-------------CFSVALENG--FRIFN WD 2 101-138 DLL--------------QK--ET---ITLKFM--SSIKDVFLSRIHI-------------VVVL---ENT-IEIFQ WD 3 199-253 LSQIKYNSQNPKESALLPT--SI---IK-AHK--NPIKLVRLNRQGT-------------MVATCSVQGTLIRIFS WD 4 257-297 -GT--------------LI--KE---FR-RGVDKADIYEMSFSPNGS-------------KLAVLSNKQT-LHIFQ WD 5 298-340 IFE--------------TT--NTETNTP-DHS--RANGSSHPLKNYI-------------PKGLWRPKYL-DSVWS WD 6 350-395 --------------------------IF-DAH--RNDNSGDVTHDNEFYKDRCRIGWCQDSNNREQDDSL-VLVWQ WD40 Template TGE--------------CL--RT---LS-GHT--SGVTSVSFSPDGK-------------RLVSGSEDGT-IKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |