WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: Y110A7A.9a [Caenorhabditis elegans] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = Y110A7A.9a (Len=1019) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (Y110A7A.9a, Len=1019): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MPHTVPGCLHPSNGRNAIDWNDGGLIAYGSHNLIFVVDVMQYKRIQSIEHHQSAINIIRWSPARSPFADLSPSSLQPIIA 80
WD40-rpts (________________WD 1________________) (_______________________WD 2___________
Query 81 SSDISGHIIISNSVTAAQINKFSHPNSSVLQMFWYPWKDISRDFLLVLHSSSTVVLWNTVTGEKVWNHTYSHVPLFDFTV 160
WD40-rpts ___________) (___________________WD 3__________________) (_________________W
Query 161 DPFSLRNVIFSSSGNKVLVCSDIALSEPASSGHLFQITSEDNSTTSTNTNIYLITYHKAYRNTVFIAINSGLYLVNPELM 240
WD40-rpts D 4_________________) (________________WD 5________________) (
Query 241 CCLARVPIESNIVSWMPTGRRDALFAVHSNGATTFRVTKFEPTEEKKPNASFSMDKVCQTECVRPMNNQRIVAAALCPTT 320
WD40-rpts _________________WD 6_________________) (__________________WD 7_
Query 321 QSTVSVLYQSGKLAFWQLSNGRLPLLYRSSFIEDVLEFNDNLSTKSIGELSLHQLSQMGSLSSGVTCVRMRPMDELTKVE 400
WD40-rpts ________________) (_______
Query 401 NDPFANTALGTLHLAAVGTNSGTVHLVDVFTSQIYRDFCVQPSLVKCLEWGGVYSLVTAGYNHSLSASQIVRNDVFITDI 480
WD40-rpts ________WD 8_______________) (_____________________WD 9_____________________)(
Query 481 RTGLAKRIRPETDESPITIIRVSYYHCYLAIAFQREPLEIWDLKGLRMLRKMSRSCPLIIDMAWSNKHHSIKTTESTLQS 560
WD40-rpts ___________________WD10__________________) (_________________WD11_____________
Query 561 VYRENLVLLDSENRIYHITLRGLHVRDGKIVNTQWKSASAQICAMAWKDDILAVGDVEGNLVVWDLGRRQSRQVRDVSHS 640
WD40-rpts ___) (____________________WD12___________________) (___________
Query 641 RVHRMTFSRLAGDHTMAVLHSKEVTLWDTEAMTRIQSIRMDSSKLCLDADLCGLSPLVLSNDNTLRFVVSNTKNQPILEK 720
WD40-rpts ______WD13_________________)
Query 721 EIPFLYKDDSMNNVRKHLEGSGSRKSSEDRSSADKSEDSTLEDAIAWIDQNQRPQIETLEADLSKLQRERVVRRFIGDHF 800
WD40-rpts
Query 801 SADLLSIVIDRFAHSETEHLAPNLQMFWENGAFKERESRVISVACAAEQAMERRLVEQAVVVGGSAKERVTDRLIVSADL 880
WD40-rpts
Query 881 RYASIKAALLVSCQDNEKAKSLIKLIATNLIASDLIEDGVELLFLVGAGGDACKYLQSQKLWTKSIVYAKMGLDDPTEVE 960
WD40-rpts
Query 961 SKWIGHLADEAKQIRVLAEASRRNWPDVVEATSSADAILARLILRTTTSTPPLSSCPTP 1019
WD40-rpts
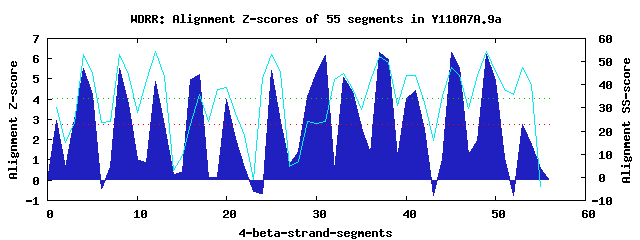 13 WD40-repeats found in QUERY=Y110A7A.9a (Len=1019): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-38 2.96 236.82 0.03933 15.0% 30.07 WD 2: Seg 4 42-92 5.52 342.57 0.00150 17.5% 53.17 WD 3: Seg 8 96-138 5.51 342.19 0.00152 17.5% 53.08 WD 4: Seg 12 142-181 4.90 317.06 0.00332 12.5% 54.53 WD 5: Seg 17 199-236 5.20 329.60 0.00225 2.5% 36.01 WD 6: Seg 20 240-279 3.98 279.26 0.01069 17.5% 38.61 WD 7: Seg 25 297-337 5.41 338.03 0.00173 7.5% 53.01 WD 8: Seg 31 393-428 6.16 368.95 0.00066 12.5% 24.09 WD 9: Seg 33 432-479 5.08 324.39 0.00264 15.0% 44.77 WD10: Seg 37 480-522 6.32 375.80 0.00054 17.5% 52.38 WD11: Seg 41 526-564 4.44 298.20 0.00595 10.0% 43.83 WD12: Seg 45 581-625 6.30 375.01 0.00055 20.0% 47.56 WD13: Seg 49 629-668 6.23 372.01 0.00060 17.5% 54.41Multiple sequence alignment of 13 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-38 MPHTVPG-----CLHPSNGRNA---------I-DWND----GG--------------LIAYGSHNLIFVVD WD 2 42-92 -YKRIQS-----IEHHQSAINI---------I-RWSP----ARSPFADLSPSSLQPIIASSDISGHIIISN WD 3 96-138 -AAQINK-----FSHPNSSVLQ---------M-FWYPWKDISR------------DFLLVLHSSSTVVLWN WD 4 142-181 -GEKVWN-----HTYSHVPLFD---------F-TVDP---FSL------------RNVIFSSSGNKVLVCS WD 5 199-236 SED--NS-----TTSTNTNIYL---------I-TYHK----AY------------RNTVFIAINSGLYLVN WD 6 240-279 -MCCLAR-----VPIESNIVSW---------MPTGRR----DA------------LFAVHSNGATTFRVTK WD 7 297-337 VCQTECV-----RPMNNQRIVA---------A-ALCP---TTQ------------STVSVLYQSGKLAFWQ WD 8 393-428 ----MDE-----LTKVENDPFA---------N-TALG----TL------------HLAAVGTNSGTVHLVD WD 9 432-479 -SQIYRD-----FCVQPSLVKCLEWGGVYSLV-TAGY----NH------------SLSASQIVRNDVFITD WD10 480-522 IRTGLAK--RIRPETDESPITI---------I-RVSY----YH------------CYLAIAFQREPLEIWD WD11 526-564 -LRMLRK-----MSRSCPLIID---------M-AWSN----KH------------HSIKTTESTLQSVYRE WD12 581-625 RGLHVRDGKIVNTQWKSASAQI---------C-AMAW----KD------------DILAVGDVEGNLVVWD WD13 629-668 -RQSRQV-----RDVSHSRVHR---------M-TFSR---LAG------------DHTMAVLHSKEVTLWD WD40 Template TGECLRT-----LSGHTSGVTS---------V-SFSP----DG------------KRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |