WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: T05C3.2 [Caenorhabditis elegans] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = T05C3.2 (Len=1733) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (T05C3.2, Len=1733): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MYSSSISRSDVHKKRSKSTSTSQLTRSSKKSSIESKFLIPNHLQKKENGNSRSTTGLDMLLSQNNEQKQVVERRDPYTQE 80
WD40-rpts
Query 81 IFDRVFSGKLSNIPKPQSKLVRVFTSSTFTDTTVERNALMEEIYPKLKEYCRETYGLDFQVVDMRWGVRDESTDDHMTTK 160
WD40-rpts
Query 161 LCLSEIANCQRLSVGANFVVFLCQKYGYRPIPSEILSTELEMLKRTLREEHEDISLLDNWYIEDSNSVPPMFVLQQISSI 240
WD40-rpts
Query 241 LVNFNNKRIPKLQEQDAKQWWECEAKMQQLLRKGARSCYEKGFLDREAMHNYFMSVTEREVVHGILKVKNPNEHCLCYIR 320
WD40-rpts
Query 321 HINNIALSQMKTASKFVDIAHNHVNSEAQKLLANLRDERVPAKLAMQNIRRSTVEWVGRDGIDVQYHAEYIRQFCTDFYK 400
WD40-rpts
Query 401 SITSMVDNAMEKHARFRDQLFTEVLTHLTTGITVSNMFFGRNEQLDCAKQYVLGNSTLPIILQGENGCGKTSLMAKIATK 480
WD40-rpts
Query 481 IREWYDNSCEPVVLLRFLGTSPDSSNIAPLLTSVCDQIASNYDESLKNSSPTELSKLFQHFKKITNLATREKPLVIIFDS 560
WD40-rpts
Query 561 LDLLSRIDGAHELLWFPPSLPPFVKLFASLTPGGSLIYSKMQRLIEDKRQYLMVPSLGKDLGFQVVREWLSDKGRTLSER 640
WD40-rpts
Query 641 QWNTVSKALDKCTLPLFVKLIYATVARWKSYSRPQETILFSSLQESINVLFHRTESQHGKLLVSHALSYISAARSGISDS 720
WD40-rpts
Query 721 EVEDLISLDDKVLDDIFQYHLPPVRRIPPLLWSRIRSDLPGYLSERAADGVIVLNWYHQQFRQVAIERYFKNVNHLETCH 800
WD40-rpts
Query 801 SAMAEYFLGVWGGVPKPYQYTEMQKQRFGVTENEGLADRKVPKQPNVFSSKDGNHRYNTRKLNELPYHLLRSGRVDELLT 880
WD40-rpts
Query 881 LCLFDYEFLHAKVSSFPLQSLIADYEDAIQNVRDVEISRQLSLAVDALRLSASILSRNPSMLAFELLGRLLPLVATNKYV 960
WD40-rpts
Query 961 SKLLIKCDKEGPQHNSFVPAHHCFHAPGGPLKFSLEEHQFAVFGMQLTSDRKLLVSTSTQIIVWDVSTGDIARVVNPNID 1040
WD40-rpts (______________WD 1______________) (___________
Query 1041 GVFFGLAVSENDKFAAAYTNNNQVIVASLVTGEFTAIDPEPFVTQMELQAIRFVGNNNVLMWSKSHYMIYTVNGNLVSQG 1120
WD40-rpts ______WD 2_________________) (___
Query 1121 SESEGEHNNIVHIFYRDALNMMFLLWTGERDEWKLVLKGSINDEKTQSKGKIANFSCEAAITFSDNYFRKGFACIVHKSS 1200
WD40-rpts _______________WD 3__________________) (___________________WD 4________________
Query 1201 LDPDGDHNFALVQIELNDSKYTTSEIIHEGLPNRINDIKIFERNCGSSQSQSQNPWIVGVMIDSFLLYRENCEFKPIFLK 1280
WD40-rpts __) (________________WD 5________________)
Query 1281 LPLNVRNIPIRPRHTTTAVTFASHDTVFVAGVRKHLFLWNVSTTELLRSLDAHFGRILNLDSVSHQGQNILISSSLDHTI 1360
WD40-rpts (________________WD 6________________) (__________________WD 7______________
Query 1361 KIWNMENIFEKSFPVSTMEQSIEKISVAKDNPTLAAVQTRKNIGIWDIKSHRYIASTIVAIESDQLLLWDLRTQSVIHSV 1440
WD40-rpts ___) (_________________WD 8________________) (_________
Query 1441 HAPNVFQIFYMNKEALIGVIYRQVDSAEQKIARLTIYSITDFSVQYSFEYPCRLFKECAVMRDGITGVVITLFKGHDSLL 1520
WD40-rpts ________WD 9_________________) (__________________WD10______
Query 1521 VFDAVEKTQKIKFRPRQSKKQKDVMINKIIAMPSNSNQIIVVESDNKSSVWDIRVRKFHRSLTQFNGVVSNDGKLGLFAP 1600
WD40-rpts ____________) (______________WD11_____________) (______________WD12______
Query 1601 AKGGLFVIDMRSGQVAKTLIGNVTEGVNDVTCSFSPNAQYVFYNHSAQDGSLLTVVLNDPMVKEETLNKIALLPCRRHLA 1680
WD40-rpts ________) (__________________WD13__________________)
Query 1681 EFQHIHVPEEAELDTFDLRNLGAVTAAVTRFKSLLDNKKGGGVTKKSHVCSIM 1733
WD40-rpts
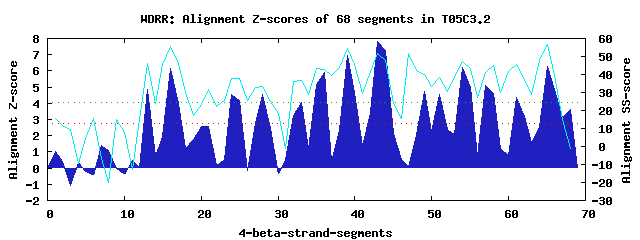 13 WD40-repeats found in QUERY=T05C3.2 (Len=1733): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 13 992-1025 4.84 314.76 0.00356 32.5% 46.21 WD 2: Seg 16 1029-1068 6.13 367.73 0.00069 17.5% 55.69 WD 3: Seg 24 1117-1158 4.55 302.43 0.00522 10.0% 38.03 WD 4: Seg 28 1161-1203 4.57 303.26 0.00509 10.0% 33.30 WD 5: Seg 33 1243-1280 4.08 283.37 0.00941 7.5% 36.40 WD 6: Seg 36 1283-1320 5.88 357.69 0.00094 15.0% 42.58 WD 7: Seg 39 1324-1364 6.93 400.86 0.00025 40.0% 54.33 WD 8: Seg 43 1369-1407 7.79 436.36 8.2e-05 20.0% 51.89 WD 9: Seg 49 1431-1470 4.75 310.88 0.00402 7.5% 40.11 WD10: Seg 54 1492-1533 6.21 371.29 0.00062 17.5% 47.19 WD11: Seg 58 1540-1572 4.63 305.82 0.00470 10.0% 44.81 WD12: Seg 61 1576-1609 4.38 295.70 0.00643 22.5% 45.67 WD13: Seg 65 1613-1654 6.28 374.09 0.00057 20.0% 56.80Multiple sequence alignment of 13 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 992-1025 ------KFSLEEHQF---AV-F---GM-QLTSDRKL-LV---STSTQ--IIVWD WD 2 1029-1068 -G--DIARVVNPNID---GVFF---GL-AVSENDKF-AA---AYTNNNQVIVAS WD 3 1117-1158 -V--SQGSESEGEHN---NI-V---HI-FYRDALNM-MFLLWTGERDEWKLVLK WD 4 1161-1203 IN--DEKTQSKGKIA---NF-SCEAAI-TFSDNYFR-KG---FACIVHKSSLDP WD 5 1243-1280 ----RNCGSSQSQSQ---NP-W---IV-GVMIDSFL-LY---RENCEFKPIFLK WD 6 1283-1320 -L--NVRNIPIRPRH---TT-T---AV-TFASHDTV-FV---AGV-RKHLFLWN WD 7 1324-1364 -T--ELLRSLDAHFG---RI-L---NLDSVSHQGQNILI---SSSLDHTIKIWN WD 8 1369-1407 -F--EKSFPVSTMEQ---SI-E---KI-SVAKDNPT-LA---AVQTRKNIGIWD WD 9 1431-1470 LR--TQSVIHSVHAP---NV-F---QI-FYMNKEAL-IG---VIYRQVDSAEQK WD10 1492-1533 CRLFKECAVMRDGIT---GV-V---IT-LFKGHDSL-LV---FDAVEKTQKIKF WD11 1540-1572 ---------KQKDVM---IN-K---II-AMPSNSNQ-II---VVESDNKSSVWD WD12 1576-1609 -R--KFHRSLTQF------------NG-VVSNDGKL-GL---FAPAKGGLFVID WD13 1613-1654 -G--QVAKTLIGNVTEGVND-V---TC-SFSPNAQY-VF---YNHSAQDGSLLT WD40 Template TG--ECLRTLSGHTS---GV-T---SV-SFSPDGKR-LV---SGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |