WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: T01G1.1d [Caenorhabditis elegans] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = T01G1.1d (Len=337) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (T01G1.1d, Len=337): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MRGNVITRTHTLEGHARGVLSVDVNEKLMVTGSKDRTAKLWDIEACREIRTLGIHPNNVHLVKFVPFSNYVFTFSMFEAR 80
WD40-rpts (________________WD 1________________) (________________WD 2______________
Query 81 AWDYRTPECICVKVLNSSGQVNEGDSIDVSQVMPRQNTIPFLETLITAADVDPTGQLLFTSFSAYVRVWNLREWKPLGRL 160
WD40-rpts __)(_____________WD 3_____________)(_______________WD 4______________) (______
Query 161 NAASHSPKSEVSCLRTTMTPEGSILAYTGSRDHYVKEYEVGLGTGVIESKCEFTPPHYDNVTAVLPLNGHLYTASKDVNI 240
WD40-rpts ______________WD 5____________________) (__________________WD 6_______________
Query 241 MKFSLKDGKREHLELRAHQQYIQSLTGFGPKGKELLVSACKDGTIRFWDVGSSSRMKLVEEYSKAHQEGINDMCSTKSML 320
WD40-rpts ___)(____________________WD 7___________________)(___________________WD 8_______
Query 321 FTASGDSTVGFWKSNAV 337
WD40-rpts ____________)
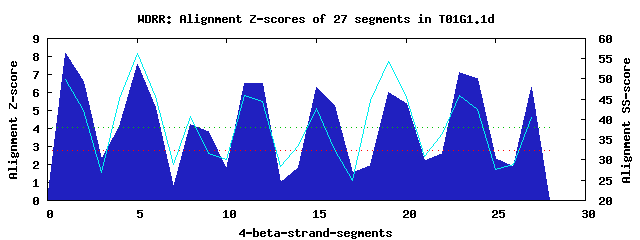 8 WD40-repeats found in QUERY=T01G1.1d (Len=337): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 5-42 8.18 452.42 5.0e-05 42.5% 49.76 WD 2: Seg 5 46-83 7.57 427.31 0.00011 27.5% 56.24 WD 3: Seg 9 84-115 3.83 272.83 0.01303 7.5% 31.66 WD 4: Seg 12 116-150 6.50 383.26 0.00043 20.0% 44.42 WD 5: Seg 15 154-199 6.26 373.05 0.00058 25.0% 42.82 WD 6: Seg 19 203-244 6.00 362.35 0.00081 15.0% 54.38 WD 7: Seg 23 245-289 7.10 408.13 0.00020 42.5% 45.81 WD 8: Seg 27 290-333 6.30 374.90 0.00055 20.0% 40.47Multiple sequence alignment of 8 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 5-42 VIT------RTH---TLEGHARG---VLS------VD-V--NEKL-MVT-GSKDRTAKLWD WD 2 46-83 -CR------EIR---TLGIHPNN---VHL------VK-FVPFSNY-VFT-FSMF-EARAWD WD 3 84-115 -----------------YRTPEC---ICV------KV-LNSSGQV-NEG-DSIDVSQVMPR WD 4 116-150 ----------QN---TIPFLETL---ITA------AD-VDPTGQL-LFT--SFSAYVRVWN WD 5 154-199 -WK------PLG---RLNAASHS---PKSEVSCLRTT-MTPEGSI-LAYTGSRDHYVKEYE WD 6 203-244 -GT------GVI---ESKCEFTPPHYDNV------TA-VLPLNGH-LYT-ASKDVNIMKFS WD 7 245-289 LKD------GKREHLELRAHQQY---IQS------LTGFGPKGKELLVS-ACKDGTIRFWD WD 8 290-333 VGSSSRMKLVEE---YSKAHQEG---IND------MC-STKSM---LFT-ASGDSTVGFWK WD40 Template TGE------CLR---TLSGHTSG---VTS------VS-FSPDGKR-LVS-GSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |