WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: R01H10.3d [Caenorhabditis elegans] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = R01H10.3d (Len=495) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (R01H10.3d, Len=495): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAQIVRQSKFRHVFCKPVKHESCMSDIRVTEITWDSLFCDVNPKFIAFINRGAGGPFMVIPVNKIGRVDKDYPFVDAHKA 80
WD40-rpts (______
Query 81 PCLEVAWSPFNDNVIASCSEDTTCKVWVIPDRGLNRNLSEPAVELTGHQKRVNTIAWHPVANNVLLTAGGENVMFMWNVG 160
WD40-rpts _________WD 1______________) (_________________WD 2_________________)
Query 161 TGEALLEISGHPDQIWSINFNFDGSQFVTTCKDKKIRILDSHTGEVVHEGMGHEGVKPQRAIFVKDGLILSTGFTKRSER 240
WD40-rpts (_________________WD 3________________) (_________________WD 4_______________
Query 241 LYALRAPEDLSTPIVEEELDTSNGVVFPFYDEDSGLVYLVGKGDCAIRYYEVNNDAPYVHYINTYTTNEPQRAVGFQSKR 320
WD40-rpts __) (_____________________WD 5____________________) (__________________WD 6_____
Query 321 GMSSEENEINRIYKLTTKGVVDILQFFVPRKSDLFQHDLYPDTRSTIPALTAEEFMEGKNAAPNRQPVNAAAAAAAAKPK 400
WD40-rpts _____________)
Query 401 VQVAKKANILSTLAPTAAESVPTQSYSERPPSSQQPSPRPSASPRPRPVVDDDMGIVTMREAPPSRPASSRASRTEIPPK 480
WD40-rpts
Query 481 EESKVDPMKPMRTTL 495
WD40-rpts
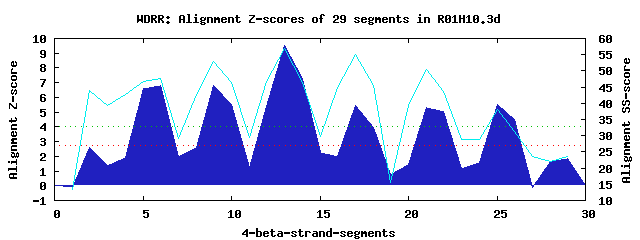 6 WD40-repeats found in QUERY=R01H10.3d (Len=495): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 6 74-108 6.80 395.44 0.00029 30.0% 47.65 WD 2: Seg 9 119-158 6.80 395.43 0.00029 20.0% 52.92 WD 3: Seg 13 162-200 9.53 508.18 8.8e-06 35.0% 56.82 WD 4: Seg 17 204-243 5.45 339.68 0.00164 22.5% 54.99 WD 5: Seg 21 245-291 5.31 334.02 0.00196 15.0% 50.44 WD 6: Seg 25 293-334 5.50 341.82 0.00154 12.5% 38.13Multiple sequence alignment of 6 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 74-108 ---------------FVDAHKA-PCLEVAWSPFNDNVIASCSEDTTCKVWV WD 2 119-158 -SE-------P--AVELTGHQK-RVNTIAWHPVANNVLLTAGGENVMFMWN WD 3 162-200 -GE-------A--LLEISGHPD-QIWSINFNF-DGSQFVTTCKDKKIRILD WD 4 204-243 -GE-------V--VHEGMGHEGVKPQRAIFVK-DGLILSTGFTKRSERLYA WD 5 245-291 RAPEDLSTPIV--EEELDTSNG-VVFPFYDED-SGLVYLVGKGDCAIRYYE WD 6 293-334 NND-------APYVHYINTYTT-NEPQRAVGF-QSKRGMSSEENEINRIYK WD40 Template TGE-------C--LRTLSGHTS-GVTSVSFSP-DGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |