WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: K10D2.1b [Caenorhabditis elegans] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = K10D2.1b (Len=669) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (K10D2.1b, Len=669): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MGDVYVTSPTFVGHDTGSILAIDCHPSGKKFITCGQKARTSNGLVVVWNAEPVLDKKKASNENVPKLLFQVESQSQSNSC 80
WD40-rpts (__________________WD 1__________________) (________________
Query 81 RWSPDGKRFAFGSDDSSVSVWEYVGLINSMGSITGGAQNVERYKECCVLRGHSMEVLTVEWSPNGKYLASGSIDYRIIIY 160
WD40-rpts _WD 2________________) (_________________WD 3________________
Query 161 NARKLPDRITVLNDIQLPVKGLSWDPIGKYLASLEGDKKLRFWATDSWQCVKSVTEPFESNIEETMLTRLDWSPDGKYLM 240
WD40-rpts ) (_________________WD 4________________) (______________________WD 5______
Query 241 TPAAVRSGKPLIKLIQRQTWKSDQFLAGHHKGTTCVRAMPRLIEANLKNGKRMQLTCAAVGSRDKSISIWVFPGTLKPLF 320
WD40-rpts _______________) (_______________________WD 6_______________________) (_____
Query 321 VINNIFNHTVMDFAWCGRNLLACSQDGTVKVIHLSESVIGEMISNEAMSDLCYQIYSIRPPRYELTDKEEDESQDSFNLS 400
WD40-rpts ____________WD 7________________) (__________________WD 8__________________)(_
Query 401 DLSSSANNASFVTCPEDVLIKRKKLVAAQQSSDIQLTKSMEDNSKENESKNSEKTMMEERNKQIDVRKDGKRRIQPVFCG 480
WD40-rpts _________WD 9__________) (_____________
Query 481 TTTADPMTSLSSEGKKTVAPPPAKRKALAPAVPAKKGEVDLEDSSDSDDDDEEEEEDMEISDIESVRNKKRKVPATTSQP 560
WD40-rpts ______WD10___________________)
Query 561 MLALDLKKPALRPMEPKSLKTQEGTVLMEAPEQQPKLSQHVVDRKGMFVEIDNRWKHGGVETTQIKLIKKKQQTAENEDD 640
WD40-rpts
Query 641 MMDGERRVNHECLWMAVLGSPVIIVAANK 669
WD40-rpts
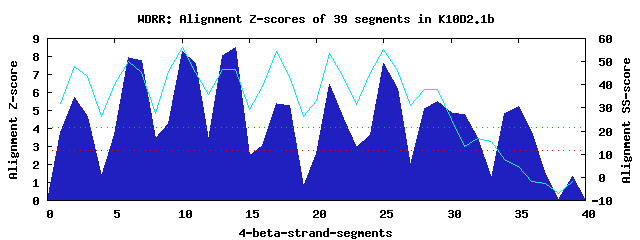 10 WD40-repeats found in QUERY=K10D2.1b (Len=669): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 8-49 5.71 350.68 0.00117 25.0% 47.71 WD 2: Seg 6 64-102 7.95 442.98 6.7e-05 35.0% 50.14 WD 3: Seg 10 123-161 8.26 455.83 4.5e-05 37.5% 55.93 WD 4: Seg 14 166-204 8.49 465.40 3.3e-05 25.0% 46.39 WD 5: Seg 17 208-256 5.40 337.67 0.00175 25.0% 54.48 WD 6: Seg 21 260-311 6.46 381.59 0.00045 30.0% 53.47 WD 7: Seg 25 315-353 7.63 429.78 0.00010 25.0% 55.08 WD 8: Seg 29 357-398 5.50 341.66 0.00155 15.0% 38.00 WD 9: Seg 32 399-424 3.49 258.87 0.02003 17.5% 16.87 WD10: Seg 35 467-510 5.23 330.75 0.00217 20.0% 4.49Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 8-49 ----SP----TFV------GHDTG--SILAIDCHP-------------SGKKFITCGQKARTSNG----LVVVWN WD 2 64-102 -VPKLL----FQV------ESQSQ---SNSCRWSP-------------DGKRFAFG-----SDDS----SVSVWE WD 3 123-161 -YKECC----VLR------GHSME---VLTVEWSP-------------NGKYLASG-----SIDY----RIIIYN WD 4 166-204 -PDRIT----VLN------DIQLP---VKGLSWDP-------------IGKYLASL-----EGDK----KLRFWA WD 5 208-256 -WQCVK----SVTEPFESNIEETM---LTRLDWSP-------------DGKYLMTP-----AAVRSGKPLIKLIQ WD 6 260-311 -WKSDQ----FLA------GHHKG---TTCVRAMPRLIEANLKNGKRMQLTCAAVG-----SRDK----SISIWV WD 7 315-353 -TLKPL----FVI------NNIFN---HTVMDFAW-------------CGRNLLAC-----SQDG----TVKVIH WD 8 357-398 -SVIGE----MIS------NEAMSDLCYQIYSIRP-------------PRYELTDK-----EEDE----SQDSFN WD 9 399-424 ---------------------------LSDLSSSA-------------NNASFVTC-----PEDV----LIKRKK WD10 467-510 RKDGKRRIQPVFC------GTTTA---DPMTSLSS-------------EGKKTVAP-----PPAK----RKALAP WD40 Template TGECLR----TLS------GHTSG---VTSVSFSP-------------DGKRLVSG-----SEDG----TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |