WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: JC8.2 [Caenorhabditis elegans] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = JC8.2 (Len=467) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (JC8.2, Len=467): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MDGVLMVSDLSWIQRGVAKEIPDKIKLDDEKLKELIEGAIPESTDNETELDDDEESDNEKLKGLRGRGDVEMKDEESEFE 80
WD40-rpts
Query 81 KKYMKGYNENIKDEPEGQDGMKGIAMYSTNKDDPYVTEQVDSDEEEEKDEIMVRKDDNMVAVAKIDKGDFTLECYVYNEA 160
WD40-rpts
Query 161 DSDWYCHHDYILDAPPLCIEPVQHDPGNEETGKGNLLAVGTMNSEIHIWDLDIMNTATPFLTLGKKERKVKGAARKKRDN 240
WD40-rpts (___________________WD 1__________________) (_________________
Query 241 SAQGHTDAVISLAWNRITTHVLASGGADKTVVLWDLDEAKPAQIIPDQGGEIQTMKWHPNESTFLLLGTMKGQVNVVDCR 320
WD40-rpts _______WD 2_______________________) (_________________WD 3_________________)(_
Query 321 ESSGNASAAWKFDGQIEKVIWNHFNPFTAFCSSDDGRLRHLDMRKPGECLWEGVAHDGPIGGLTLSALTKGLLVTVGEDE 400
WD40-rpts __________________WD 4___________________) (_________________WD 5____________
Query 401 MMNVWKVEDTNGGIEKVHSEKLTIGELHCAQFNPDVAAVLSVGGTAADLIRIIDLTKYEPVVKAFSE 467
WD40-rpts _____)(_____________________WD 6_____________________)
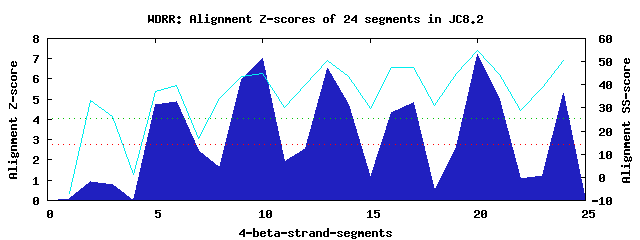 6 WD40-repeats found in QUERY=JC8.2 (Len=467): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 6 168-210 4.90 316.91 0.00333 22.5% 39.65 WD 2: Seg 10 223-275 7.00 403.99 0.00022 32.5% 44.82 WD 3: Seg 13 279-318 6.52 383.84 0.00042 15.0% 50.65 WD 4: Seg 17 319-362 4.86 315.40 0.00349 15.0% 47.60 WD 5: Seg 20 367-406 7.22 412.95 0.00017 32.5% 54.72 WD 6: Seg 24 407-454 5.27 332.43 0.00206 20.0% 50.51Multiple sequence alignment of 6 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 168-210 HD------------------YILDAPPLCIEPVQHDPGNEETGKG-N-L-LAV--GTMNSEIHIWD WD 2 223-275 LGKKERKVKGAARKKR---DNSAQGHTDAVISLAWNR------ITTH-V-LAS--GGADKTVVLWD WD 3 279-318 -A------------KP---AQIIPDQGGEIQTMKWHP------NE-S-TFLLL--GTMKGQVNVVD WD 4 319-362 CR------------ESSGNASAAWKFDGQIEKVIWNH-----FNP-F-T-AFC--SSDDGRLRHLD WD 5 367-406 -G------------EC---LWEGVAHDGPIGGLTLSA------LT-KGL-LVT--VGEDEMMNVWK WD 6 407-454 VE------DTNGGIEK---VHSEKLTIGELHCAQFNP------DV-A-A-VLSVGGTAADLIRIID WD40 Template TG------------EC---LRTLSGHTSGVTSVSFSP------DG-K-R-LVS--GSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |