WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: FBpp0087322 [Drosophila melanogaster] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = FBpp0087322 (Len=949) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (FBpp0087322, Len=949): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MKFSYKFSNLLGTIYRNGNLVFTPDGNSVISPVGNRITVYDLKNNKSRTLSLESRYNYTNIALSPDGSLLIAVNEKGEAQ 80
WD40-rpts (_______________WD 1______________) (_________________WD 2______________
Query 81 LISMMSCTVIHRHKFQTGVQCICFSPNGAFFAVAVQNVVLIFCAPGEITGEYNPFVLRRRFLGGFDDVTWLDWSSDSRLL 160
WD40-rpts __)(__________________WD 3_________________)(______________________WD 4_________
Query 161 AIGCRDSSTKIVAINYTKNFRTYNLSGHTDAIVACFFEANSLHLNTISRNGQLCLWECSLAPSDLEDAERPKIAPPQRKK 240
WD40-rpts ____________)(___________________WD 5___________________)(__________________WD 6
Query 241 EKKAVSDNESEDDVENVVEKDALKEESTKEAEDVNHIKNEDETKSHPFFYKKITRHYLADEPRKEQRDVQLTAANYNTRT 320
WD40-rpts _________________) (__________________WD 7__
Query 321 KVLVVAFSSGAFYLYELPDVNMIHSLSISDYPISAALFNCTGDWVALASREIGQLLVWEWQSEQYIMKQQGHSSEMTCIA 400
WD40-rpts _______________) (_________________WD 8_________________) (_________________
Query 401 YSSDGQFIATGGEDSKVKLWNTQSSFCFVTFSEHTSGVTGIQFSRNKKFLVSSSLDGTVRAFDVNRYRNFRTFTSPNPAQ 480
WD40-rpts WD 9________________) (_________________WD10________________)(________________
Query 481 FSCVAVDYSGELVVAGGQDVFDIFLWSVKTGKLLEIISGHEGPVVSIAFSPVATSSTLVSGSWDKTVKIWNCLESNSEHE 560
WD40-rpts ___WD11___________________) (__________________WD12_________________) (_____
Query 561 TIDAVSDVTNVTFSPSGEEIAVATLSGNITIFDIKSAGQVTTIEGRNDLSAGRLETDIITARKNAQANYFSTIEYSADGE 640
WD40-rpts ____________WD13________________) (______________WD1
Query 641 CILAAGKSANICIYHVREAILLKKFEITQNHSLDGLNDFISRKHLSEFGNMALVEEREELEGGRVAIRLPGVQRGDMSSR 720
WD40-rpts 4_____________) (____________________WD15___________________) (_____
Query 721 RFQQEVRVFSVKFSPTGQAFAAAGTEGLCIYALDKGVVFDPFDLSLEVTPKAVHESLKQQNYTKALVMSLKLNEPNLIAL 800
WD40-rpts ___________WD16________________)
Query 801 VLERVPYKDVELVCADLSPEFAQRLLQQLARQLQSTPHIEFYLQWSCCLLTKHGNQDGVFQHTGLLALHEVLSRKYEMLN 880
WD40-rpts
Query 881 KICDYNKYTLKVLLDRADKLEQKNEGKPNATEEDSDEEMLLIRSKDDENGQAQFSDEEESDSGSEEEMA 949
WD40-rpts
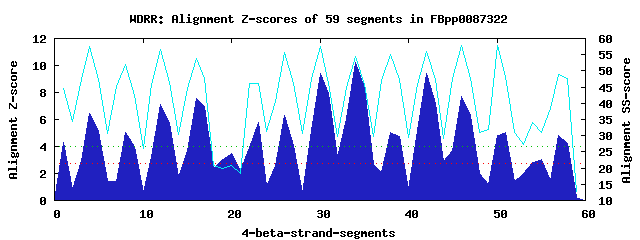 16 WD40-repeats found in QUERY=FBpp0087322 (Len=949): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 7-41 4.29 291.84 0.00724 25.0% 44.51 WD 2: Seg 4 45-83 6.44 380.62 0.00046 20.0% 57.68 WD 3: Seg 8 84-124 5.07 324.26 0.00265 17.5% 52.07 WD 4: Seg 12 125-173 7.14 409.65 0.00019 30.0% 56.63 WD 5: Seg 16 174-217 7.52 425.27 0.00012 25.0% 53.75 WD 6: Seg 20 218-258 3.51 259.62 0.01958 2.5% 20.93 WD 7: Seg 23 296-336 5.78 353.34 0.00108 12.5% 46.10 WD 8: Seg 26 340-379 6.26 373.15 0.00058 20.0% 55.81 WD 9: Seg 30 383-421 9.39 502.71 1.0e-05 30.0% 57.53 WD10: Seg 34 425-463 10.25 538.05 3.5e-06 50.0% 54.53 WD11: Seg 38 464-507 5.05 323.27 0.00274 20.0% 54.97 WD12: Seg 42 511-551 9.44 504.73 9.8e-06 47.5% 56.05 WD13: Seg 46 555-593 7.69 432.14 9.3e-05 27.5% 57.94 WD14: Seg 51 623-655 5.01 321.54 0.00289 10.0% 48.25 WD15: Seg 55 667-711 3.07 241.51 0.03410 12.5% 31.13 WD16: Seg 57 715-752 4.83 314.24 0.00362 22.5% 48.81Multiple sequence alignment of 16 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 7-41 ------------------FSNLLGTIYRNGNLVFTP--DGNSVISPVGN--------RITVYD WD 2 45-83 -N-K---------S----RTLSLESRYNYTNIALSP--DGSLLIAVNEK-G------EAQLIS WD 3 84-124 MM-S---------C---TVIHRHKFQTGVQCICFSP--NGAFFAVAVQN-V------VLIFCA WD 4 125-173 PG-EITGEYNPFVL----RRRFLGGFDDVTWLDWSS--DSRLLAIGCRD-S------STKIVA WD 5 174-217 IN-Y---------TKNFRTYNLSGHTDAIVACFFEA--NSLHLNTISRN-G------QLCLWE WD 6 218-258 CS-L---------A---PSDLEDAERPKIAPPQRKK--EKKAVSDNESE-D------DVENVV WD 7 296-336 HYLA---------D----EPRKEQRDVQLTAANYNT--RTKVLVVAFSS-G------AFYLYE WD 8 340-379 -V-N---------M----IHSLSISDYPISAALFNC--TGDWVALASREIG------QLLVWE WD 9 383-421 -E-Q---------Y----IMKQQGHSSEMTCIAYSS--DGQFIATGGED-S------KVKLWN WD10 425-463 -S-F---------C----FVTFSEHTSGVTGIQFSR--NKKFLVSSSLD-G------TVRAFD WD11 464-507 VN-R------YRNF----RTFTSPNPAQFSCVAVDY--SGELVVAGGQDVF------DIFLWS WD12 511-551 -G-K---------L----LEIISGHEGPVVSIAFSPVATSSTLVSGSWD-K------TVKIWN WD13 555-593 -S-N---------S----EHETIDAVSDVTNVTFSP--SGEEIAVATLS-G------NITIFD WD14 623-655 ---------------------KNAQANYFSTIEYSA--DGECILAAGKS-A------NICIYH WD15 667-711 IT-Q--------------NHSLDGLNDFISRKHLSE--FGNMALVEERE-ELEGGRVAIRLPG WD16 715-752 -G-D---------M----SSRRFQQEVRVFSVKFSP--TGQAFAAAGTE-G-------LCIYA WD40 Template TG-E---------C----LRTLSGHTSGVTSVSFSP--DGKRLVSGSED-G------TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |