WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSP00000418287 [Homo sapiens] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSP00000418287 (Len=339) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSP00000418287, Len=339): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MKDSLVLLGRVPAHPDSRCWFLAWNPAGTLLASCGGDRRIRIWGTEGDSWICKSVLSEGHQRTVRKVAWSPCGNYLASAS 80
WD40-rpts (__________________WD 1_________________)(____________________WD 2___________
Query 81 FDATTCIWKKNQDDFECVTTLEGHENEVKSVAWAPSGNLLATCSRDKSVWVWEVDEEDEYECVSVLNSHTQDVKHVVWHP 160
WD40-rpts ________) (_________________WD 3________________) (_________________WD
Query 161 SQELLASASYDDTVKLYREEEDDWVCCATLEGHESTVWSLAFDPSGQRLASCSDDRTVRIWRQYLPGNEQGVACSGSDPS 240
WD40-rpts 4________________)(___________________WD 5___________________)
Query 241 WKCICTLSGFHSRTIYDIAWCQLTGALATACGDDAIRVFQEDPNSDPQQPTFSLTAHLHQAHSQDVNCVAWNPKEPGLLA 320
WD40-rpts (_________________WD 6_________________)
Query 321 SCSDDGEVAFWKYQRPEGL 339
WD40-rpts
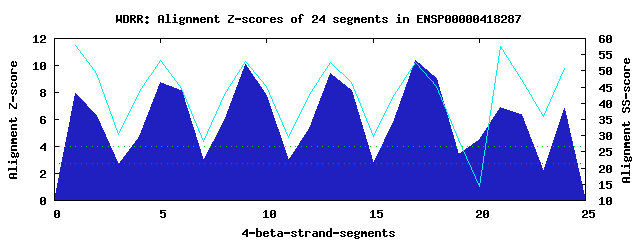 6 WD40-repeats found in QUERY=ENSP00000418287 (Len=339): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 4-44 7.92 441.99 6.9e-05 25.0% 57.83 WD 2: Seg 5 45-89 8.74 475.66 2.4e-05 35.0% 53.12 WD 3: Seg 9 95-133 10.08 530.95 4.3e-06 40.0% 52.92 WD 4: Seg 13 140-178 9.42 503.93 1.0e-05 35.0% 52.59 WD 5: Seg 17 179-222 10.37 543.22 3.0e-06 47.5% 52.62 WD 6: Seg 21 241-280 6.86 397.95 0.00027 25.0% 57.57Multiple sequence alignment of 6 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 4-44 SLV-----LLGRVPA-HPDSRCWFLAWNPAGTLLASCGGDRRIRIWG WD 2 45-89 TEGDSWICKSVLSEG-HQ-RTVRKVAWSPCGNYLASASFDATTCIWK WD 3 95-133 -FE-----CVTTLEG-HE-NEVKSVAWAPSGNLLATCSRDKSVWVWE WD 4 140-178 -YE-----CVSVLNS-HT-QDVKHVVWHPSQELLASASYDDTVKLYR WD 5 179-222 EEE-DDWVCCATLEG-HE-STVWSLAFDPSGQRLASCSDDRTVRIWR WD 6 241-280 -WK-----CICTLSGFHS-RTIYDIAWCQLTGALATACGDDAIRVFQ WD40 Template TGE-----CLRTLSG-HT-SGVTSVSFSPDGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |