WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSP00000395972 [Homo sapiens] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSP00000395972 (Len=174) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSP00000395972, Len=174): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MGGGGSALRVCADHRGGINWLSLSPDGQRLLTGSEDGTARLWSTADGQCCALLQGRPSHSLPDLFAAAHRPQTADPSSCA 80
WD40-rpts (_________________WD 1_________________) (_________________WD 2____________
Query 81 RRPLSRSPAPAHPGTLAEERLSGPSIWLLSARQGRVPCGLFVANCALLLAFPVDDSHSNSRRLLRASGAEKCAVVTLCYP 160
WD40-rpts ____) (___________________WD 3__________________)
Query 161 TYRRGNRLGDMKAM 174
WD40-rpts
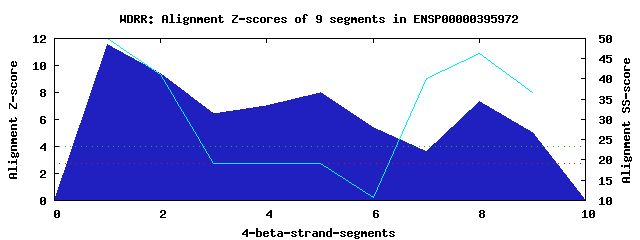 3 WD40-repeats found in QUERY=ENSP00000395972 (Len=174): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 4-43 11.55 591.79 6.6e-07 47.5% 49.92 WD 2: Seg 5 47-85 8.01 445.43 6.2e-05 15.0% 19.19 WD 3: Seg 8 103-145 7.32 416.96 0.00015 17.5% 46.41Multiple sequence alignment of 3 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 4-43 GGSALRVCADHRGGINWLSLSPDGQRLLT----GSEDGTARLWS WD 2 47-85 -GQCCALLQGRPSHSLPDLFAAAHRPQTA----DPSSCARRPLS WD 3 103-145 -GPSIWLLSARQGRVPCGLFVANCALLLAFPVDDSHSNSRRLLR WD40 Template TGECLRTLSGHTSGVTSVSFSPDGKRLVS----GSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |