WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSP00000386746 [Homo sapiens] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSP00000386746 (Len=288) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSP00000386746, Len=288): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MSAFEKPQIIAHIQKGFNYTVFDCKWVPCSAKFVTMGNFARGTGVIQLYEIQHGDLKLLREIEKAKPIKCGTFGATSLQQ 80
WD40-rpts (___________________WD 1___________________)(____________________WD 2_____
Query 81 RYLATGDFGGNLHIWNLEAPEMPVYSVKGHKEIINAIDGIGGLGIGEGAPEIVTGSRDGTVKVWDPRQKDDPVANMEPVQ 160
WD40-rpts _______________) (________________WD 3________________) (____
Query 161 GENKRDCWTVAFGNAYNQEERVVCAGYDNGDIKLFDLRNMALRWETNIKNGVCSLEFDRKDISMNKLVATSLEGKFHVFD 240
WD40-rpts ______________WD 4_________________) (__________________WD 5__________________)
Query 241 MRTQHPTKGFASVSEKAHKSTVWQVRHLPQNRELFLTAGGAGGLHLWK 288
WD40-rpts (_________________WD 6_________________)
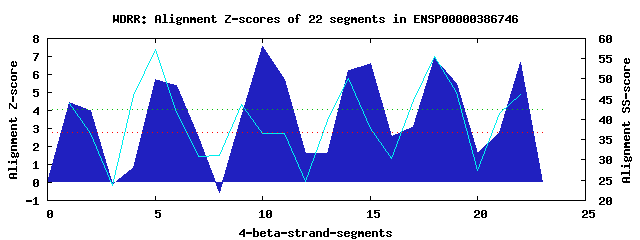 6 WD40-repeats found in QUERY=ENSP00000386746 (Len=288): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 7-50 4.46 299.01 0.00580 17.5% 43.96 WD 2: Seg 5 51-96 5.72 350.95 0.00116 17.5% 57.27 WD 3: Seg 10 108-145 7.56 426.96 0.00011 30.0% 36.66 WD 4: Seg 15 156-196 6.61 387.51 0.00037 12.5% 37.87 WD 5: Seg 18 199-240 6.91 400.17 0.00025 30.0% 55.51 WD 6: Seg 22 249-288 6.65 389.52 0.00035 22.5% 46.08Multiple sequence alignment of 6 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 7-50 PQI---IA-HIQKGFNYTVFDCKW---VPCSA---KFV-TMGNFARGTGVIQLYE WD 2 51-96 IQHGDLKL-LREIEKAKPIKCGTFGATSL-QQ---RYL-ATGDFG---GNLHIWN WD 3 108-145 KGH------KEIINAIDGIGGLGI---GE-GA---PEI-VTGSRD---GTVKVWD WD 4 156-196 MEP---VQGENKRDCWTVAFGNAY---NQ-EE---RVV-CAGYDN---GDIKLFD WD 5 199-240 -NM---AL-RWETNIKNGVCSLEF---DR-KDISMNKL-VATSLE---GKFHVFD WD 6 249-288 -GF---AS-VSEKAHKSTVWQVRH---LP-QN---RELFLTAGGA---GGLHLWK WD40 Template TGE---CL-RTLSGHTSGVTSVSF---SP-DG---KRL-VSGSED---GTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |