WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSP00000308450 [Homo sapiens] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSP00000308450 (Len=499) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSP00000308450, Len=499): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAQFAFESDLHSLLQLDAPIPNAPPARWQRKAKEAAGPAPSPMRAANRSHSAGRTPGRTPGKSSSKVQTTPSKPGGDRYI 80
WD40-rpts
Query 81 PHRSAAQMEVASFLLSKENQPENSQTPTKKEHQKAWALNLNGFDVEEAKILRLSGKPQNAPEGYQNRLKVLYSQKATPGS 160
WD40-rpts
Query 161 SRKTCRYIPSLPDRILDAPEIRNDYYLNLVDWSSGNVLAVALDNSVYLWSASSGDILQLLQMEQPGEYISSVAWIKEGNY 240
WD40-rpts (________________WD 1________________)(___________________WD 2______
Query 241 LAVGTSSAEVQLWDVQQQKRLRNMTSHSARVGSLSWNSYILSSGSRSGHIHHHDVRVAEHHVATLSGHSQEVCGLRWAPD 320
WD40-rpts _____________) (________________WD 3_______________) (_________________WD 4
Query 321 GRHLASGGNDNLVNVWPSAPGEGGWVPLQTFTQHQGAVKAVAWCPWQSNVLATGGGTSDRHIRIWNVCSGACLSAVDAHS 400
WD40-rpts ________________) (__________________WD 5__________________) (__________
Query 401 QVCSILWSPHYKELISGHGFAQNQLVIWKYPTMAKVAELKGHTSRVLSLTMSPDGATVASAAADETLRLWRCFELDPARR 480
WD40-rpts _______WD 6_________________) (_________________WD 7________________)
Query 481 REREKASAAKSSLIHQGIR 499
WD40-rpts
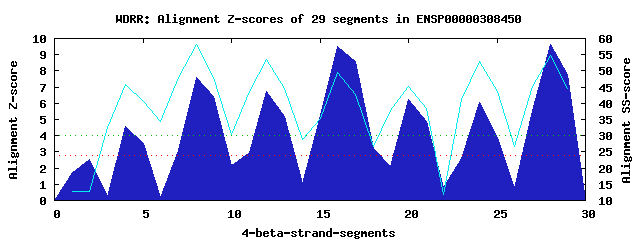 7 WD40-repeats found in QUERY=ENSP00000308450 (Len=499): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 4 173-210 4.56 302.85 0.00515 12.5% 45.78 WD 2: Seg 8 211-254 7.60 428.53 0.00010 20.0% 58.07 WD 3: Seg 12 258-294 6.70 391.56 0.00033 32.5% 53.49 WD 4: Seg 16 299-337 9.53 508.45 8.7e-06 37.5% 49.64 WD 5: Seg 20 345-386 6.21 371.35 0.00062 25.0% 45.07 WD 6: Seg 24 390-429 6.06 364.88 0.00075 35.0% 52.95 WD 7: Seg 28 433-471 9.65 513.35 7.5e-06 37.5% 54.76Multiple sequence alignment of 7 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 173-210 -DR----ILDAPEIRNDYYLNLVDWSSGNVLAV---AL---DNSVYLWS WD 2 211-254 ASSGDILQLLQMEQPGEYISSVAWIKEGNYLAV---GTS--SAEVQLWD WD 3 258-294 -QK----RLRNMTSHSARVGSLSW--NSYILSS---GSR--SGHIHHHD WD 4 299-337 -EH----HVATLSGHSQEVCGLRWAPDGRHLAS---GGN--DNLVNVWP WD 5 345-386 -WV----PLQTFTQHQGAVKAVAWCPWQSNVLATGGGTS--DRHIRIWN WD 6 390-429 -GA----CLSAVDAH-SQVCSILWSPHYKELIS---GHGFAQNQLVIWK WD 7 433-471 -MA----KVAELKGHTSRVLSLTMSPDGATVAS---AAA--DETLRLWR WD40 Template TGE----CLRTLSGHTSGVTSVSFSPDGKRLVS---GSE--DGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |