WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000124131 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000124131 (Len=782) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000124131, Len=782): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MYPQGRHPAPHQPGQPGFKFTVAESCDRIKDEFQFLQAQYHSLKVEYDKLANEKTEMQRHYVMYYEMSYGLNIEMHKQTE 80
WD40-rpts
Query 81 IAKRLNTILAQIMPFLSQEHQQQVAQAVERAKQVTMTELNAIIGVRGLPNLPLTQQQLQAQHLSHATHGPPVQLPPHPSG 160
WD40-rpts
Query 161 LQPPGIPPVTGSSSGLLALGALGSQAHLAVKDEKNHHELDHRERESSTNNSVSPSESLRASEKHRGSADYSMEAKKRKAE 240
WD40-rpts
Query 241 EKDSLSRYDSDGDKSDDLVVDVSNEDPATPRVSPAHSPPENGLDKARGLKKDAPTSPASVASSSSTPSSKTKDLGHNDKS 320
WD40-rpts (_________________WD 1_________________)
Query 321 STPGLKSNTPTPRNDAPTPGTSTTPGLRSMPGKPPGMDPIGIMASALRTPITLTSSYPAPFAMMSHHEMNGSLTSPSAYA 400
WD40-rpts
Query 401 GLHNIPSQMSAAAAAAAAAYGRSPMVSFGAVGFDPHPPMRATGLPSSLASIPGGKPAYSFHVSADGQMQPVPFPHDALAG 480
WD40-rpts (_______________WD 2______________)
Query 481 PGIPRHARQINTLSHGEVVCAVTISNPTRHVYTGGKGCVKIWDISQPGSKSPISQLDCLNRDNYIRSCKLLPDGRTLIVG 560
WD40-rpts (________________WD 3________________)(_____________________WD 4___________
Query 561 GEASTLTIWDLASPTPRIKAELTSSAPACYALAISPDAKVCFSCCSDGNIAVWDLHNQTLVRQFQGHTDGASCIDISHDG 640
WD40-rpts _________) (_________________WD 5________________) (_________________WD 6_
Query 641 TKLWTGGLDNTVRSWDLREGRQLQQHDFTSQIFSLGYCPTGEWLAVGMESSNVEVLHHTKPDKYQLHLHESCVLSLKFAY 720
WD40-rpts _______________)(__________________WD 7_________________) (_________________WD
Query 721 CGKWFVSTGKDNLLNAWRTPYGASIFQSKESSSVLSCDISADDKYIVTGSGDKKATVYEVIY 782
WD40-rpts 8________________)(__________________WD 9_________________)
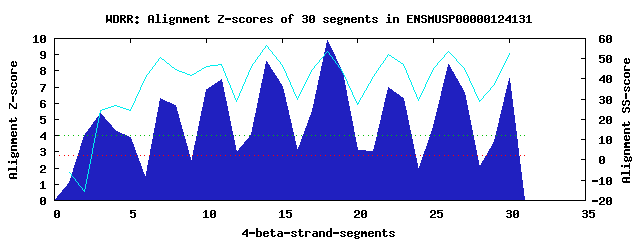 9 WD40-repeats found in QUERY=ENSMUSP00000124131 (Len=782): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 243-282 3.99 279.51 0.01061 12.5% -15.58 WD 2: Seg 3 419-453 5.36 336.16 0.00183 10.0% 24.60 WD 3: Seg 7 486-523 6.30 374.94 0.00055 22.5% 50.42 WD 4: Seg 11 524-570 7.45 422.20 0.00013 25.0% 47.15 WD 5: Seg 14 576-614 8.57 468.67 3.0e-05 30.0% 56.31 WD 6: Seg 18 618-656 9.87 522.57 5.6e-06 35.0% 53.80 WD 7: Seg 22 657-697 6.97 402.65 0.00023 25.0% 51.87 WD 8: Seg 26 700-738 8.40 461.81 3.7e-05 30.0% 53.67 WD 9: Seg 30 739-779 7.56 426.80 0.00011 32.5% 52.78Multiple sequence alignment of 9 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 243-282 DSL-------SRYDSD-GDKSDDLVVDVSNEDPATPRVSPAHSPPENG WD 2 419-453 ------------AYGR-SPMVSFGAVGFDPHPPMRATGLPSSLASIPG WD 3 486-523 -HA-------RQINTL-SHGEVVCAVTISNPTRHVYTGG-KGCVKIWD WD 4 524-570 ISQPGSKSPISQLDCL-NRDNYIRSCKLLPDGRTLIVGGEASTLTIWD WD 5 576-614 -PR-------IKAELT-SSAPACYALAISPDAKVCFSCCSDGNIAVWD WD 6 618-656 -QT-------LVRQFQ-GHTDGASCIDISHDGTKLWTGGLDNTVRSWD WD 7 657-697 LRE-------GRQLQQHDFTSQIFSLGYCPTGEWLAVGMESSNVEVLH WD 8 700-738 -KP-------DKYQLH-LHESCVLSLKFAYCGKWFVSTGKDNLLNAWR WD 9 739-779 TPY------GASIFQS-KESSSVLSCDISADDKYIVTGSGDKKATVYE WD40 Template TGE-------CLRTLS-GHTSGVTSVSFSPDGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |