WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000121654 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000121654 (Len=281) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000121654, Len=281): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 XIWGAYDGKFEKTISGHKLGISDVAWSSDSNLLVSASDDKTLKIWDVSSGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSF 80
WD40-rpts (_________________WD 1_________________) (_________________WD 2_________
Query 81 DESVRIWDVKTGKCLKTLPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTLIVFVTSHEKRSPLRTPCDDD 160
WD40-rpts _______) (_________________WD 3________________) (_________
Query 161 NPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCLKTYTGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEI 240
WD40-rpts ________WD 4________________) (__________________WD 5__________________) (__
Query 241 VQKLQGHTDVVISTACHPTENIIASAALENDKTIKLWKSDC 281
WD40-rpts ________________WD 6_________________)
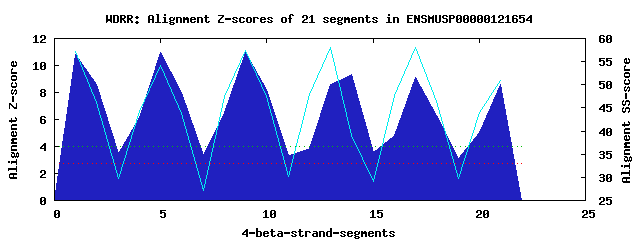 6 WD40-repeats found in QUERY=ENSMUSP00000121654 (Len=281): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 7-46 10.82 561.48 1.7e-06 45.0% 57.28 WD 2: Seg 5 50-88 10.94 566.48 1.4e-06 42.5% 54.08 WD 3: Seg 9 92-130 10.98 568.20 1.4e-06 45.0% 57.48 WD 4: Seg 14 151-189 9.31 499.35 1.2e-05 30.0% 38.98 WD 5: Seg 17 193-234 9.10 490.38 1.5e-05 37.5% 58.02 WD 6: Seg 21 238-278 8.60 469.73 2.9e-05 35.0% 50.99Multiple sequence alignment of 6 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 7-46 DGKFEKTISGHKLGISDVAWS---SDSNLLVSAS--DDKTLKIWD WD 2 50-88 -GKCLKTLKGHSNYVFCCNFN---PQSNLIVSGS--FDESVRIWD WD 3 92-130 -GKCLKTLPAHSDPVSAVHFN---RDGSLIVSSS--YDGLCRIWD WD 4 151-189 -SPLRTPCDDDNPPVSFVKFS---PNGKYILAAT--LDNTLKLWD WD 5 193-234 -GKCLKTYTGHKNEKYCIFANFSVTGGKWIVSGS--EDNLVYIWN WD 6 238-278 -KEIVQKLQGHTDVVISTACH---PTENIIASAALENDKTIKLWK WD40 Template TGECLRTLSGHTSGVTSVSFS---PDGKRLVSGS--EDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |