WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000116772 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000116772 (Len=1076) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000116772, Len=1076): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MMAALAAGGYTRSDTIEKLSSVMAGVPARRNQSSPPPAPPLCLRRRTRLAAAPEDTVQNRVTLEKVLGITAQNSSGLTCD 80
WD40-rpts (_____________________WD 1___
Query 81 PGTGHVAYLAGCVVVVLNPKENKQQHIFNTTRKSLSALAFSPDGKYIVTGENGHRPAVRIWDVEEKTQVAEMLGHKYGVA 160
WD40-rpts _________________) (__________________WD 2_________________) (______________
Query 161 CVAFSPNMKHIVSMGYQHDMVLNVWDWKKDIVVASNKVSCRVIALSFSEDSSYFVTVGNRHVRFWFLEASTEAKVTSTVP 240
WD40-rpts ____WD 3_________________) (________________WD 4________________)
Query 241 LVGRSGILGELHNNIFCGVACGRGRMAGNTFCVSYSGLLCQFNEKRVLDKWINLKVSLSSCLCVSDELIFCGCTDGIVRI 320
WD40-rpts (_______________WD 5______________) (_________________WD 6________________
Query 321 FQAHSLLYLTNLPKPHYLGVDVAHGLDSSFLFHRKAEAVYPDTVALTFDPVHQWLSCVYKDHSIYIWDVKDIDEVSKIWS 400
WD40-rpts _) (____________________WD 7___________________)(___________
Query 401 ELFHSSFVWNVEVYPEFEDQRACLPSGTFLTCSSDNTIRFWNLDSASDTRWQKNIFSDSLLKVVYVENDIQHLQDLSHFP 480
WD40-rpts _____________WD 8________________________)
Query 481 DRGSENGTPMDMKAGVRVMQVSPDGQHLASGDRSGNLRIHELHFMDELIKVEAHDAEVLCLEYSKPETGVTLLASASRDR 560
WD40-rpts (_________________WD 9________________) (__________________WD10_____________
Query 561 LIHVLNVEKNYNLEQTLDDHSSSITAIKFAGTRDVQMISCGADKSIYFRSAQQASDGLHFVRTHHVAEKTTLYDMDIDIT 640
WD40-rpts _____) (_________________WD11________________)(_____________________WD12____
Query 641 QKYVAVACQDRNVRVYNTVSGKQKKCYKGSQGDEGSLLKVHVDPSGTFLATSCSDKSISLIDFYSGECVAKMFGHSEIVT 720
WD40-rpts ________________) (_________________WD13________________) (______________
Query 721 GMKFTYDCRHLITVSGDSCVFIWHLGPEITTCMKQHLLEINHQEQQQQPKDQKWSGPPSQETYASTPSEIRSLSPGEQTE 800
WD40-rpts ___WD14________________)(_______________________WD15_______________________)
Query 801 DEMEEECEPEELLKTPSKDSLDPDPRCLLTNGKLPLWAKRLLGDDDVADSSAFHAKRSYQPHGRWAERAEQEPLKTILDA 880
WD40-rpts (___________________WD16__________________)
Query 881 WSLDSYFTPMKPENLQDSVLDSVEPQNLAGLLSECSLGNGHTSPGEGLVSYLLHPELGSPKEDNRGHPSYLPLQREATEA 960
WD40-rpts (_________________WD17________________) (_________________WD18_
Query 961 SELILCSPEAEVSLTGMHREYYEEETEAGPEDQQGDTYLRVSSVSSKDQSPPEDSGESEAELECSFAAAHSSAPQTDPGP 1040
WD40-rpts _______________)
Query 1041 HLTMTAASSAVTQSADKSSPPCLPSRVPKYRRAFPA 1076
WD40-rpts
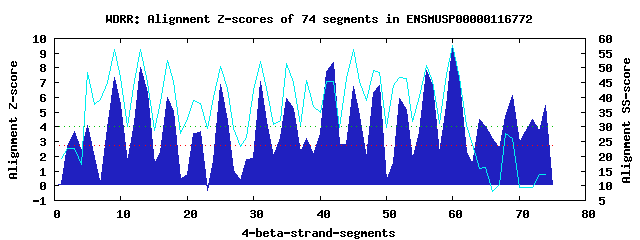 18 WD40-repeats found in QUERY=ENSMUSP00000116772 (Len=1076): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 5 52-98 4.10 284.06 0.00922 10.0% 48.36 WD 2: Seg 9 102-142 7.32 417.18 0.00015 25.0% 56.41 WD 3: Seg 13 146-186 8.02 445.99 6.1e-05 37.5% 56.19 WD 4: Seg 17 189-226 5.97 361.38 0.00084 17.5% 52.47 WD 5: Seg 22 247-281 3.68 266.83 0.01568 5.0% 38.04 WD 6: Seg 25 283-322 6.86 397.88 0.00027 10.0% 50.47 WD 7: Seg 31 344-388 7.09 407.59 0.00020 22.5% 52.15 WD 8: Seg 35 389-442 5.95 360.59 0.00086 25.0% 51.59 WD 9: Seg 42 483-521 8.38 460.99 3.8e-05 30.0% 45.32 WD10: Seg 45 525-566 6.72 392.05 0.00032 25.0% 56.32 WD11: Seg 49 572-610 6.83 396.76 0.00028 22.5% 48.47 WD12: Seg 52 611-657 5.92 359.09 0.00090 10.0% 46.91 WD13: Seg 56 664-702 7.82 437.53 7.9e-05 22.5% 50.87 WD14: Seg 60 706-744 9.49 506.84 9.2e-06 32.5% 57.63 WD15: Seg 64 745-796 4.48 299.82 0.00566 15.0% 16.01 WD16: Seg 69 837-879 6.11 367.18 0.00070 12.5% 26.22 WD17: Seg 72 883-921 4.50 300.36 0.00557 12.5% 9.55 WD18: Seg 74 938-976 5.47 340.44 0.00161 15.0% 13.90Multiple sequence alignment of 18 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 52-98 AP-EDTVQNRVTL-----EK---VLG-------ITAQNSS-----------GLTCDPGT---G---------HVAYLA---G--CVVVVLN WD 2 102-142 -N-K--------Q-----QH---IFN-------TTRKSLS-----------ALAFSPDG---K---------YIVTGE--NGHRPAVRIWD WD 3 146-186 -K-T--------Q-----VA---EML-------GHKYGVA-----------CVAFSPNM---K---------HIVSMGYQHD--MVLNVWD WD 4 189-226 -K-D--------I-----VV---ASN-------KVSCRVI-----------ALSFSEDS---S---------YFVTVG---N--RHVRFWF WD 5 247-281 -------------------I---LGE-------LHNNIFC-----------GVACGRGR---M---------AGNTFC--VS--YSGLLCQ WD 6 283-322 NE-K--------R-----VL---DKW-------INLKVSL-----------SSCLCVSD---E---------LIFCGC--TD--GIVRIFQ WD 7 344-388 HG-L---DSSFLF-----HR---KAE-------AVYPDTV-----------ALTFDPVH---Q---------WLSCVY--KD--HSIYIWD WD 8 389-442 VK-D--------IDEVSKIW---SEL-------FHSSFVW-----------NVEVYPEF---EDQRACLPSGTFLTCS--SD--NTIRFWN WD 9 483-521 -G-S--------E-----NG---TPM-------DMKAGVR-----------VMQVSPDG---Q---------HLASGD--RS--GNLRIHE WD10 525-566 -M-D--------E-----LI---KVE-------AHDAEVL-----------CLEYSKPETGVT---------LLASAS--RD--RLIHVLN WD11 572-610 ---N--------L-----EQ---TLD-------DHSSSIT-----------AIKFAGTR---D--------VQMISCG--AD--KSIYFRS WD12 611-657 AQ-Q--------A-----SD---GLHFVRTHHVAEKTTLY-----------DMDIDITQ---K---------YVAVAC--QD--RNVRVYN WD13 664-702 -K-K--------C-----YK---GSQ-------GDEGSLL-----------KVHVDPSG---T---------FLATSC--SD--KSISLID WD14 706-744 -G-E--------C-----VA---KMF-------GHSEIVT-----------GMKFTYDC---R---------HLITVS--GD--SCVFIWH WD15 745-796 LGPE--------I-----TT---CMK-------QHLLEINHQEQQQQPKDQKWSGPPSQ---E---------TYASTP--SE--IRSLSPG WD16 837-879 WA-K--------R-----LLGDDDVA-------DSSAFHA-----------KRSYQPHG---R---------WAERAE--QE--PLKTILD WD17 883-921 -L-D--------S-----YF---TPM-------KPENLQD-----------SVLDSVEP---Q---------NLAGLL--SE--CSLGNGH WD18 938-976 -G-S--------P-----KE---DNR-------GHPSYLP-----------LQREATEA---S---------ELILCS--PE--AEVSLTG WD40 Template TG-E--------C-----LR---TLS-------GHTSGVT-----------SVSFSPDG---K---------RLVSGS--ED--GTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |