WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000112491 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000112491 (Len=841) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000112491, Len=841): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MATGGRAWGGDRARAGAAGAGGGCGGAMAERGPAFCGLYDTSSLLQYCNDDNLSGTSGMEVDDRVSALEQRLQLQEDELA 80
WD40-rpts
Query 81 VLKAALADALRRLRACEEQGAALRARSTPKGRAPPRLGTTASVCQLLKGLPTRTPLNGSGPPRRVGGYATSPSSPKKEAS 160
WD40-rpts (___________________WD 1_________
Query 161 SGRSSRRYLSPERLASVRREDPRSRTTSSSSNCSAKKEGKTKEVIFSMEEGSVKMFLRGRPVPMLIPDELAPTYSLDTRS 240
WD40-rpts _________) (_____________________WD 2____________________)
Query 241 ELPSSRLKLDWVYGYRGRDCRANLYLLPTGEVVYFVASVAVLYSVEEQRQRHYLGHNDDIKCLAVHPDMVTIATGQVAGT 320
WD40-rpts (______________________WD 3_______
Query 321 TKEGKPLPPHVRVWDSVSLSTLHVLGLGVFDRAVCCVAFSKSNGGNLLCAVDESNDHVLSVWDWAKESKVVDSKCSNEAV 400
WD40-rpts ______________)(_____________________WD 4_____________________) (_____________
Query 401 LVATFHPTDPSLLITCGKSHIYFWSLEGGSLSKRQGLFEKHEKPKYVLCVTFLEGGDVVTGDSGGNLYVWGKGGNRITQE 480
WD40-rpts ____WD 5________________) (________________WD 6________________)(________
Query 481 VQGAHDGGVFALCALRDGTLVSGGGRDRRVVLWGSDYSKVQEVEVPEDFGPVRTVAEGRGDTLYVGTTRNSILLGSVHTG 560
WD40-rpts ___________WD 7__________________) (_________________WD 8________________) (_
Query 561 FSLLVQGHVEELWGLATHPSRAQFVTCGQDKLVHLWSSETHQPVWSRSIEDPARSAGFHPSGSVLAVGTVTGRWLLLDTE 640
WD40-rpts ________________WD 9________________) (_________________WD10________________)
Query 641 THDLVAIHTDGNEQISVVSFSPDGAYLAVGSHDNLVYVYTVDQGGRKVSRLGKCSGHSSFITHLDWAQDSTCFVTNSGDY 720
WD40-rpts (_________________WD11________________) (_________________WD12___________
Query 721 EILYWDPVTCKQITSADTVRNVEWATATCVLGFGVFGIWPEGADGTDINAVARSHDGKLLVSADDFGKVHLFSYPCCQPR 800
WD40-rpts _____) (_____________________WD13_____________________) (
Query 801 ALSHKYGGHSSHVTNVAFLWDDSMALTTGGKDTSVLQWRVA 841
WD40-rpts _________________WD14_________________)
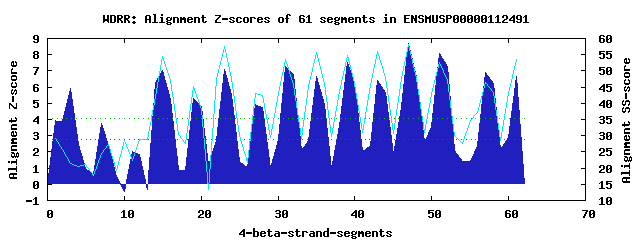 14 WD40-repeats found in QUERY=ENSMUSP00000112491 (Len=841): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 3 128-170 5.90 358.52 0.00092 22.5% 21.38 WD 2: Seg 7 189-235 3.76 270.14 0.01416 10.0% 24.50 WD 3: Seg 15 287-335 7.00 403.61 0.00023 27.5% 54.46 WD 4: Seg 19 336-383 5.32 334.49 0.00193 27.5% 45.19 WD 5: Seg 23 387-425 7.10 407.84 0.00020 15.0% 57.53 WD 6: Seg 27 434-471 4.90 317.23 0.00330 25.0% 43.14 WD 7: Seg 31 472-514 7.25 414.11 0.00016 22.5% 53.46 WD 8: Seg 35 518-556 6.65 389.46 0.00035 12.5% 55.67 WD 9: Seg 39 559-597 7.53 425.73 0.00011 15.0% 54.76 WD10: Seg 43 600-638 6.43 380.32 0.00047 20.0% 55.84 WD11: Seg 47 642-680 8.67 472.84 2.6e-05 30.0% 58.28 WD12: Seg 51 688-726 8.10 449.09 5.5e-05 32.5% 52.63 WD13: Seg 57 746-793 6.89 399.28 0.00026 27.5% 46.47 WD14: Seg 61 800-839 6.68 390.65 0.00034 30.0% 53.41Multiple sequence alignment of 14 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 128-170 KGL---P--------TRTPLNGSG-------PPRRVGGYATS-PSSP----KKEASSGRSS----------RRYLS WD 2 189-235 SSS---N--------CSA---KKEGKTKEVIFSMEEGSVKMF-LRGR----PVPMLIPDEL----------APTYS WD 3 287-335 EQR------------QRH---YLG-------HNDDIKCLAVH-PDMV----TIATGQVAGTTKEGKPLPPHVRVWD WD 4 336-383 SVS---L----STLHVLG---LGV-------FDRAVCCVAFS-KSNGGNLLCAVDESNDHV----------LSVWD WD 5 387-425 -ES---K--------VVD---SKC-------SNEAVLVATFH-PTDP----SLLITCGKSH----------IYFWS WD 6 434-471 -RQ---G--------LFE---KHE-------KPKYVLCVTFL-EGGD-----VVTGDSGGN----------LYVWG WD 7 472-514 KGGNRIT--------QEV---QGA-------HDGGVFALCAL-RDGT----LVSGGGRDRR----------VVLWG WD 8 518-556 -SK---V--------QEV---EVP-------EDFGPVRTVAE-GRGD----TLYVGTTRNS----------ILLGS WD 9 559-597 -TG---F--------SLL---VQG-------HVEELWGLATH-PSRA----QFVTCGQDKL----------VHLWS WD10 600-638 -TH---Q--------PVW---SRS-------IEDPARSAGFH-PSGS----VLAVGTVTGR----------WLLLD WD11 642-680 -HD---L--------VAI---HTD-------GNEQISVVSFS-PDGA----YLAVGSHDNL----------VYVYT WD12 688-726 -VS---R--------LGK---CSG-------HSSFITHLDWA-QDST----CFVTNSGDYE----------ILYWD WD13 746-793 TAT---CVLGFGVFGIWP---EGA-------DGTDINAVARS-HDGK----LLVSADDFGK----------VHLFS WD14 800-839 -RA---L--------SHK---YGG-------HSSHVTNVAFLWDDSM----ALTTGGKDTS----------VLQWR WD40 Template TGE---C--------LRT---LSG-------HTSGVTSVSFS-PDGK----RLVSGSEDGT----------IKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |