WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000105078 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000105078 (Len=510) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000105078, Len=510): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MTKHTLSMLRCFHTKGVNYINFSATGKLLVSVGVDPEHTITVWRWQEGTKVASRGGHLERIFVVEFRPDSDTQFVSVGVK 80
WD40-rpts (_________________WD 1_________________) (_________________WD 2___________
Query 81 HMKFWTLAGSALLYKKGVIGSMEAAKMQTMLSVAFGANNLTFTGAINGDVYVWKEHFLIRLVAKAHTGPVFTMYTTLRDG 160
WD40-rpts _____) (________________WD 3________________)(_____________________WD 4
Query 161 LIVTGGKERPTKEGGAVKLWDQEMKRCRAFQLETGQLVECVRSVCRGKGKILVGTKDGEIIEVGEKSAASNILIDGHMEG 240
WD40-rpts ____________________)(___________________WD 5___________________) (____________
Query 241 EIWGLATHPSKDMFISASNDGTARIWDLADKKLLNKVNLGHAARCAAYSPDGEMVAIGMKNGEFVILLVNTLKVWGKKRD 320
WD40-rpts _____WD 6_________________)(__________________WD 7_________________) (________
Query 321 RKSAIQDIRISPDNRFLAVGSSEQTVDFYDLTQGTSLNRIGYCKDIPSFVIQMDFSADSKYIQVSTGAYKRQVHEVPLGK 400
WD40-rpts _________WD 8________________) (_______________WD 9_______________)
Query 401 QVTEAMVVEKITWASWTSVLGDEVIGIWPRNADKADVNCACVTHAGLNIVTGDDFGLLKLFDFPCTEKFAKHKRYFGHSA 480
WD40-rpts (_____________________WD10_____________________) (___________
Query 481 HVTNIRFSSDDKYVVSTGGDDCSVFVWRCL 510
WD40-rpts ______WD11_________________)
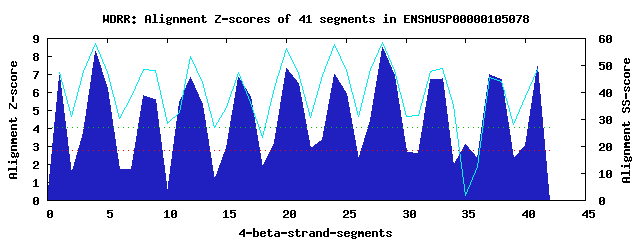 11 WD40-repeats found in QUERY=ENSMUSP00000105078 (Len=510): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 5-44 6.93 400.85 0.00025 40.0% 47.24 WD 2: Seg 4 48-86 8.30 457.68 4.2e-05 22.5% 57.96 WD 3: Seg 8 97-134 5.85 356.15 0.00099 20.0% 48.42 WD 4: Seg 12 135-181 6.85 397.59 0.00027 25.0% 53.20 WD 5: Seg 16 182-225 6.86 398.21 0.00027 17.5% 47.43 WD 6: Seg 20 228-267 7.31 416.80 0.00015 27.5% 56.46 WD 7: Seg 24 268-308 6.99 403.57 0.00023 17.5% 57.82 WD 8: Seg 28 312-350 8.52 466.81 3.2e-05 22.5% 58.67 WD 9: Seg 33 360-395 6.76 393.71 0.00031 22.5% 48.93 WD10: Seg 37 415-462 6.99 403.60 0.00023 17.5% 45.59 WD11: Seg 41 469-508 7.46 422.80 0.00012 35.0% 48.68Multiple sequence alignment of 11 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 5-44 ---------------TLSM-LRCFHTKG-VNYINFSATGKLLVS-VGVD----PEHTITVWR WD 2 48-86 -G----T-K--------VA-SRGGHLER-IFVVEFRPDSDTQFV-SVGV------KHMKFWT WD 3 97-134 -G----V-I--------GS-MEAAKMQT-MLSVAFGANNL-TFT-GAIN------GDVYVWK WD 4 135-181 EH----F-L--------IRLVAKAHTGP-VFTMYTTLRDGLIVT-GGKERPTKEGGAVKLWD WD 5 182-225 QEMKRCR-A--------FQ-LETGQLVE-CVRSVCRGKGKILVG-TKDG------EIIEVGE WD 6 228-267 -A----A-S--------NI-LIDGHMEGEIWGLATHPSKDMFIS-ASND------GTARIWD WD 7 268-308 LA----DKK--------LL-NKVNLGHA-ARCAAYSPDGEMVAI-GMKN------GEFVILL WD 8 312-350 -L----K-V--------WG-KKRDRKSA-IQDIRISPDNRFLAV-GSSE------QTVDFYD WD 9 360-395 IG----Y-C-------------KDIPSF-VIQMDFSADSKYIQV-STGA------YKRQVHE WD10 415-462 SW----T-SVLGDEVIGIW-PRNADKAD-VNCACVTHAGLNIVT-GDDF------GLLKLFD WD11 469-508 -F----A-K--------HK-RYFGHSAH-VTNIRFSSDDKYVVSTGGDD------CSVFVWR WD40 Template TG----E-C--------LR-TLSGHTSG-VTSVSFSPDGKRLVS-GSED------GTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |