WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000101195 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000101195 (Len=1155) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000101195, Len=1155): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MRKFNIRKVLDGLTAGSSSASQQQQQQQHPPGNREPEIQETLQSEHFQLCKTVRHGFPYQPSALAFDPVQKILAVGTQTG 80
WD40-rpts (____________________WD 1____________________)_________________WD 2___________
Query 81 ALRLFGRPGVECYCQHDSGAAVIQLQFLINEGALVSALADDTLHLWNLRQKRPAVLHSLKFCRERVTFCHLPFQSKWLYV 160
WD40-rpts _____) (_________________WD 3________________) (_________________WD 4______
Query 161 GTERGNIHIVNVESFTLSGYVIMWNKAIELSSKAHPGPVVHISDNPMDEGKLLIGFESGTVVLWDLKSKKADYRYTYDEA 240
WD40-rpts __________) (_________________WD 5________________) (____________
Query 241 IHSVAWHHEGKQFICSHSDGTLTIWNVRSPAKPVQTITPHGKQLKDGKKPEPCKPILKVELKTTRSGEPFIILSGGLSYD 320
WD40-rpts _____WD 6________________)
Query 321 TVGRRPCLTVMHGKSTAVLEMDYSIVDFLTLCETPYPNDFQEPYAVVVLLEKDLVLIDLAQNGYPIFENPYPLSIHESPV 400
WD40-rpts (_________________WD 7_________________)
Query 401 TCCEYFADCPVDLIPALYSVGARQKRQGYSKKEWPINGGNWGLGAQSYPEIIITGHADGSVKFWDASAITLQVLYKLKTS 480
WD40-rpts (____________________WD 8____________________)
Query 481 KVFEKSRNKDDRQNTDIVDEDPYAIQIISWCPESRMLCIAGVSAHVIIYRFSKQEVLTEVIPMLEVRLLYEINDVDTPEG 560
WD40-rpts (___________________WD 9___________________)
Query 561 EQPPPLSTPVGSSNPQPIPPQSHPSTSSSSSDGLRDNVPCLKVKNSPLKQSPGYQTELVIQLVWVGGEPPQQITSLALNS 640
WD40-rpts (______________W
Query 641 SYGLVVFGNCNGIAMVDYLQKAVLLNLSTIELYGSNDPYRREPRSPRKSRQPSGAGLCDITEGTVVPEDRCKSPTSGNLL 720
WD40-rpts D10______________) (_________________WD11_________________)
Query 721 GHCFLCTEIKCSSSPHNSDDEQKVNRWEPGRPPFNTVSHADCREICMALIVSQIPDNSFSRSRSSSVTSIDKESRETISA 800
WD40-rpts
Query 801 LHFCETLTRKADSSPSPCLWVGTTVGTAFVITLNLPPGPEQRLLQPVIVSPSGTILRLKGAILRMAFLDATGCLMSPAYE 880
WD40-rpts
Query 881 PWKEHNVAEEKDEKEKLKKRRPVSVSPSSSQEISENQYAVICSEKQAKVMSLPTQSCAYKQNITETSFVLRGDIVALSNS 960
WD40-rpts
Query 961 VCLACFCANGHIMTFSLPSLRPLLDVYYLPLTNMRIARTFCFANNGQALYLVSPTEIQRLTYSQETCENLQEMLGELFTP 1040
WD40-rpts
Query 1041 VETPEAPNRGFFKGLFGGGAQSLDREELFGESSSGKASRSLAQHIPGPGGIEGVKGAASGVVGELARARLALDERGQKLS 1120
WD40-rpts
Query 1121 DLEERTAAMMSSADSFSKHAHEMMLKYKDKKWYQF 1155
WD40-rpts
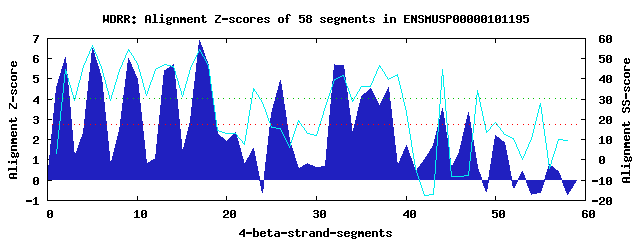 11 WD40-repeats found in QUERY=ENSMUSP00000101195 (Len=1155): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 3-48 4.57 303.35 0.00507 17.5% 3.12 WD 2: Seg 2 48-86 6.06 365.07 0.00075 20.0% 44.70 WD 3: Seg 5 89-127 6.49 382.55 0.00043 22.5% 56.44 WD 4: Seg 9 133-171 6.01 362.82 0.00080 25.0% 54.73 WD 5: Seg 14 187-225 5.71 350.69 0.00117 30.0% 46.15 WD 6: Seg 17 228-266 6.88 398.92 0.00026 20.0% 54.71 WD 7: Seg 26 341-380 4.94 318.52 0.00317 7.5% 15.47 WD 8: Seg 32 420-465 5.72 350.96 0.00116 20.0% 39.98 WD 9: Seg 38 487-530 4.58 304.04 0.00497 10.0% 39.72 WD10: Seg 44 625-658 3.53 260.46 0.01908 17.5% 44.76 WD11: Seg 47 660-699 3.37 253.89 0.02334 10.0% -7.86Multiple sequence alignment of 11 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 3-48 KFN------I--RKVLDG----LTAGSSSASQQQ-QQQQHPPGNREPEIQETLQSEHFQ WD 2 48-86 -QL------C--KTVRHG----FPYQPSALAFDP-VQKILAVGTQTG------ALRLFG WD 3 89-127 -GV------E--CYCQHD----SGAAVIQLQFLI-NEGALVSALADD------TLHLWN WD 4 133-171 -PA------V--LHSLKF----CRERVTFCHLPF-QSKWLYVGTERG------NIHIVN WD 5 187-225 ----------AIELSSKA----HPGPVVHISDNPMDEGKLLIGFESG------TVVLWD WD 6 228-266 -SK------K--ADYRYT----YDEAIHSVAWHH-EGKQFICSHSDG------TLTIWN WD 7 341-380 MDY------S--IVDFLT----LCETPYPNDFQE-PYAVVVLLEKDL------VLIDLA WD 8 420-465 VGARQKRQGY--SKKEWP----INGGNWGLGAQS-YPEIIITGHADG------SVKFWD WD 9 487-530 RNK------D--DRQNTDIVDEDPYAIQIISWCP-ESRMLCIAGVSA------HVIIYR WD10 625-658 VG--------------GE----PPQQITSLALNS-SYGLVVFGNCNG------IAMVDY WD11 660-699 QKA------V--LLNLST----IELYGSNDPYRR-EPRSPRKSRQPS------GAGLCD WD40 Template TGE------C--LRTLSG----HTSGVTSVSFSP-DGKRLVSGSEDG------TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |