WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000093960 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000093960 (Len=897) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000093960, Len=897): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MDGAAGPGEGPAHETLQTLSQRLRVQEEEMELVKAALAEALRLLRLHGSTTTLQGSGISAPTRNSSITVPPGLPPTCSPS 80
WD40-rpts
Query 81 LVTRGTQTEEELEIVPSSGPPGLSNGPPALQGGSEEPSGTQSEGGCSSSSGAGSPGPPGILRPVQPLQRSDTPRRNSSSS 160
WD40-rpts
Query 161 SSPSERPRQKLSRKAASSANLLLRSGSTESRGNKDPLSSPGGPGSRRSNYNLEGISVKMFLRGRPITMYIPSGIRSLEEL 240
WD40-rpts (_________________WD 1________________) (__________
Query 241 PSGPPPETLSLDWVYGYRGRDSRSNLFVLRSGEVVYFIACVVVLYRPGGGPGGPGGGGQRHYRGHTDCVRCLAVHPDGVR 320
WD40-rpts _______WD 2________________) (_________________WD 3___
Query 321 VASGQTAGVDKDGKPLQPVVHIWDSETLLKLQEIGLGAFERGVGALAFSAADQGAFLCVVDDSNEHMLSVWDCSRGVKLA 400
WD40-rpts _____________) (_____________________WD 4_____________________) (____
Query 401 EIKSTNDSVLAVGFSPRDSSCIVTSGKSHVHFWNWSGGTGAPGNGLLARKQGVFGKYKKPKFIPCFVFLPDGDILTGDSE 480
WD40-rpts _____________WD 5________________)(________________________WD 6_________________
Query 481 GNILTWGRSVSDSKTPGRGGAKETYTIVAQAHAHEGSIFALCLRRDGTVLSGGGRDRRLVQWGPGLVALQEAEIPEHFGA 560
WD40-rpts ______) (_________________WD 7________________)(________________
Query 561 VRAIAEGLGSELLVGTTKNALLRGDLAQGFSPVIQGHTDELWGLCTHPSQNRFLTCGHDRQLCLWDGEGHALAWSMDLKE 640
WD40-rpts __WD 8__________________)(__________________WD 9_________________) (___________
Query 641 TGLCADFHPSGAVVVVGLNTGRWLVLDTETREIVSDVTDGNEQLSVVRYSPDGLYLAIGSHDNMIYIYSVSSCGTKSSRF 720
WD40-rpts ______WD10________________) (_________________WD11________________)(__________
Query 721 GRCMGHSSFITHLDWSKDGNFIMSNSGDYEILYWDVAGGCKLLRNRYESRDREWATYTCVLGFHVYGVWPDGSDGTDINS 800
WD40-rpts __________WD12____________________) (__________________
Query 801 LCRSHNERVVAVADDFCKVHLFQYPCARAKAPSRMYSGHGSHVTSVRFTHDDSYLVSLGGKDASIFQWRVLGAGSSGPAP 880
WD40-rpts WD13__________________) (_________________WD14_________________)
Query 881 ATPSRTPSLSPASSLDV 897
WD40-rpts
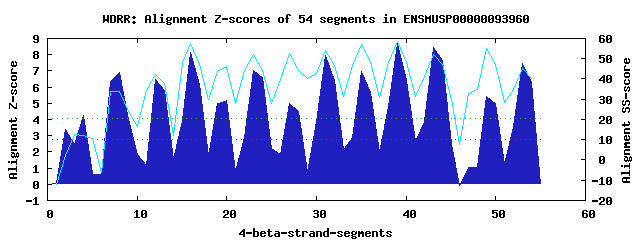 14 WD40-repeats found in QUERY=ENSMUSP00000093960 (Len=897): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 183-221 3.38 254.26 0.02308 17.5% 2.33 WD 2: Seg 4 230-268 4.23 289.40 0.00781 20.0% 12.34 WD 3: Seg 8 296-334 6.88 398.89 0.00026 35.0% 33.63 WD 4: Seg 12 345-392 6.44 380.55 0.00046 27.5% 42.39 WD 5: Seg 16 396-434 8.13 450.43 5.3e-05 22.5% 57.75 WD 6: Seg 20 435-487 5.15 327.36 0.00241 20.0% 46.11 WD 7: Seg 23 505-543 7.05 406.06 0.00021 17.5% 52.26 WD 8: Seg 27 544-585 4.99 320.87 0.00295 15.0% 52.58 WD 9: Seg 31 586-626 7.95 442.94 6.7e-05 20.0% 54.06 WD10: Seg 35 629-667 6.98 403.16 0.00023 25.0% 56.91 WD11: Seg 39 671-709 8.67 472.75 2.6e-05 27.5% 58.62 WD12: Seg 43 710-755 8.45 463.68 3.5e-05 35.0% 52.16 WD13: Seg 49 782-823 5.45 339.75 0.00164 5.0% 54.93 WD14: Seg 53 830-869 7.43 421.59 0.00013 45.0% 45.41Multiple sequence alignment of 14 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 183-221 -L--------------R-S------GS----TE--SRGNKDPLSSPGGPGSRR----SNY-NLEGISVKMFL WD 2 230-268 IP--------------S-G------IR----SL--EELPSGPPPETLSLDWVY----GYR-GR-DSRSNLFV WD 3 296-334 -G--------------G-G------QR----HY--RGHTDCVRCLAVHPDGVR----VAS-GQTAGVDKDGK WD 4 345-392 SE--------------T-L------LKLQEIGL--GAFERGVGALAFSAADQGAFLCVVD-DSNEHMLSVWD WD 5 396-434 -G--------------V-K------LA----EI--KSTNDSVLAVGFSPRDSS----CIV-TSGKSHVHFWN WD 6 435-487 WSGGTGAPGNGLLARKQ-G------VF----GK--YKKPKFIPCFVFLPDGD-----ILT-GDSEGNILTWG WD 7 505-543 -Y--------------T-I------VA----QA--HAHEGSIFALCLRRDGTV----LSG-GGRDRRLVQWG WD 8 544-585 PG-----------LVAL-Q------EA----EI--PEHFGAVRAIAEGLGSEL----LVG-TT-KNALLRGD WD 9 586-626 LA--------------QGF------SP----VI--QGHTDELWGLCTHPSQNR----FLT-CGHDRQLCLWD WD10 629-667 -G--------------H-A------LA----WS--MDLKETGLCADFHPSGAV----VVV-GLNTGRWLVLD WD11 671-709 -R--------------E-I------VS----DV--TDGNEQLSVVRYSPDGLY----LAI-GSHDNMIYIYS WD12 710-755 VS--------------S-CGTKSSRFG----RC--MGHSSFITHLDWSKDGNF----IMS-NSGDYEILYWD WD13 782-823 GF--------------H-V------YG----VWPDGSDGTDINSLCRSHNERV----VAV-ADDFCKVHLFQ WD14 830-869 -K--------------A-P------SR----MY--SGHGSHVTSVRFTHDDSY----LVSLGGKDASIFQWR WD40 Template TG--------------E-C------LR----TL--SGHTSGVTSVSFSPDGKR----LVS-GSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |