WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000082668 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000082668 (Len=532) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000082668, Len=532): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MLLSVSRILRRKNLLVTGGLDRIIRVWNPYLPGKPTGVLRGHRAPVMYVHVAFEENKVFSMSADNTVKVWDLETHGCCCT 80
WD40-rpts (___________WD 1___________) (_________________WD 2________________) (_____
Query 81 ISSKASGIKGELTACLYLPGPRALCVATGTLAFLHLKVGSVPESHLVRSHQEPVVCCRYNPAFRHVVSCCEASVVKVWDF 160
WD40-rpts ______________WD 3__________________) (_________________WD 4________________)
Query 161 ETGRQVSEFIGTHGNAGITCLTFDSSGRRLVTGGRDGCLKIWSYNNGHCLHTLQHDENQSEVCDCTYMEVNRNRCVIAVG 240
WD40-rpts (__________________WD 5_________________) (___________________WD 6__________
Query 241 WDRRINVYFDIPRDFHHFRKPQPHWQDDLNHGHKEDILCVAQCPPFLLATSSYDGEIIVWNVISGHMYCKLNTPSPMDGQ 320
WD40-rpts ________) (________________WD 7________________) (________
Query 321 DHREGPDRSVSQLAFLKTRLTKLDSASAALISNGPGGSVCFWKLFSGSGLVANFTPSREKAQVSGIVVTSGDTLAYLADQ 400
WD40-rpts _______________WD 8_______________________)(____________________WD 9____________
Query 401 HGFIHVYDIQEYGLQGTELQAPNTVTSWRAHVSMVTSLELIEGGSIVLSSSLDCTVRLWSQDGAYIGTFGQNHPWDVFTP 480
WD40-rpts _______) (_________________WD10________________)(________________WD1
Query 481 DSWGHPRVPFEILTDPRSMPTHRVLETDVPAGCLEGRQEQDHGTEEKDPCEG 532
WD40-rpts 1________________)
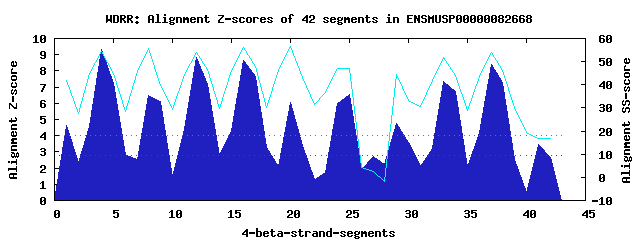 11 WD40-repeats found in QUERY=ENSMUSP00000082668 (Len=532): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-28 4.62 305.60 0.00473 17.5% 41.93 WD 2: Seg 4 33-71 9.33 500.14 1.1e-05 35.0% 54.58 WD 3: Seg 8 75-117 6.49 382.66 0.00043 20.0% 55.72 WD 4: Seg 12 121-159 8.84 479.88 2.1e-05 25.0% 54.10 WD 5: Seg 16 163-203 8.62 470.86 2.8e-05 35.0% 55.98 WD 6: Seg 20 207-249 6.07 365.55 0.00074 20.0% 56.61 WD 7: Seg 25 264-301 6.57 386.13 0.00039 27.5% 46.90 WD 8: Seg 29 312-363 4.74 310.55 0.00406 17.5% 44.34 WD 9: Seg 33 364-408 7.34 417.93 0.00014 17.5% 51.82 WD10: Seg 37 422-460 8.40 461.78 3.7e-05 27.5% 53.84 WD11: Seg 41 461-498 3.46 257.72 0.02075 7.5% 16.61Multiple sequence alignment of 11 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-28 -----------------------ML----LSVSRI--------L----RRKNLLVTGGLDRIIRVWN WD 2 33-71 -G----K-----PTGVLRGH--RAP----VMYVHV--------A----FEENKVFSMSADNTVKVWD WD 3 75-117 -H----G-----CCCTISSK--ASGIKGELTACLY--------L----PGPRALCVATGTLAFLHLK WD 4 121-159 -V----P-----ESHLVRSH--QEP----VVCCRY--------N----PAFRHVVSCCEASVVKVWD WD 5 163-203 -G----R-----QVSEFIGTHGNAG----ITCLTF--------D----SSGRRLVTGGRDGCLKIWS WD 6 207-249 -G----H-----CLHTLQHD--ENQ----SEVCDC--------TYMEVNRNRCVIAVGWDRRINVYF WD 7 264-301 -H----W-----QDDLNHGH--KED----ILCVAQ--------C----PPF-LLATSSYDGEIIVWN WD 8 312-363 NTPSPMD-----GQDHREGP--DRS----VSQLAFLKTRLTKLD----SASAALISNGPGGSVCFWK WD 9 364-408 LF----SGSGLVANFTPSRE--KAQ----VSGIVV--------T----SGDTLAYLADQHGFIHVYD WD10 422-460 -P----N-----TVTSWRAH--VSM----VTSLEL--------I----EGGSIVLSSSLDCTVRLWS WD11 461-498 ------Q-----DGAYIGTF--GQN----HPWDVF--------T----PDSWGHPRVPFEILTDPRS WD40 Template TG----E-----CLRTLSGH--TSG----VTSVSF--------S----PDGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |