WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000081567 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000081567 (Len=1223) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000081567, Len=1223): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MLPYTVNFKVSARTLTGALNAHNKAAVDWGWQGLIAYGCHSLVVVIDSNTAQTLQVLEKHKADIVKVRWARENYHHNIGS 80
WD40-rpts (_________________WD 1_________________) (______________________WD 2___
Query 81 PYCLRLASADVTGKIIVWDVAAGVAQCEIQEHVKPIQDVQWLWNQDASRDLLLAIHPPNYIVLWNADTGTKLWKKSYADN 160
WD40-rpts __________________) (___________________WD 3__________________) (___________
Query 161 ILSFSFDPFDPSHLTLLTSEGIVFISDFSPSKPPSGPGKKVYISSPHSSPAHNKLAAATGAKKALNKVKILITQEKPSAD 240
WD40-rpts ______WD 4________________) (_________________WD 5________________) (_______
Query 241 FVALNDCLQLAYLPSKRNHMLLLYPREILILDLEVNQTVGVIAIERTGVPFLQVIPCSQRDGLFCLHENGCITLRVRRSY 320
WD40-rpts __________WD 6_________________)(___________________WD 7___________________)
Query 321 NSICTTSNDEPDLDPVQELTYDLRSQCDAIRVTKTVRPFSMVCCPVNENAAALIVSDGRVMIWELKSAVCSRNARNSSGV 400
WD40-rpts (____________________WD 8___________________) (_____________
Query 401 SPLYSPVSFCGIPGGVLQNKLPDLSLDNMIGQSAIAGEEHPKGSILQEVHLKFLLTGLLSGLPSPQFAIRMCPPLTTKNI 480
WD40-rpts _______WD 9___________________) (____________________WD10
Query 481 KMYQPLLAVGTSNGSVLVYHLTSGLLHKELSVHSCEVKGIEWTSLTSFLSFAASTPNNMGLVRNELQLVDLPTGRSTAFR 560
WD40-rpts ___________________) (_____________________WD11____________________) (
Query 561 GDRGNDESPIEMIKVSHLKQYLAVVFKDKPLELWDIRTCTLLREMSKSFPAITALEWSPSHNLKSLRKKQLATREAMARQ 640
WD40-rpts _______________WD12_______________) (_________________WD13________________)
Query 641 TVVSDAELGAVESSVISLLQEAESKAELSQNISAREHFVFTDNDGQVYHLTVEGNSVKDSARIPPDGSMGSITCIAWKGD 720
WD40-rpts (_________________WD14________________) (__________________WD15____
Query 721 TLVLGDMDGNLNFWDLKARVSRGIPTHRSWVRKIRFAPGKGNQKLIAMYNDGAEVWDTKEVQMVSSLRSGRNVTFRILDV 800
WD40-rpts ______________) (_________________WD16_________________) (__________________W
Query 801 DWCTSDKVILASDDGCIRVLEMSMKSTCFRMDEQELVEPVWCPYLLVPRAALALKAFLLHQPWNGRYSLDISHIDYPENE 880
WD40-rpts D17_________________)
Query 881 EIKTLLQEQLHALSNDIKKLLLDPDFSLLQRCLLVSRLYGDESELHFWTVAAHYLHSLSQAKSGDTVVTKEGAPKDRLSN 960
WD40-rpts
Query 961 PLDICYDVLCENTYFQKFQLERVNLQEVKRSTYDHTRKCTDQLLLLGQTDRAVQLLLETSADNQHYYCDSLKACLVTTVT 1040
WD40-rpts
Query 1041 SSGPSQSTIKLVATNMIANGKLAEGVQLLCLIDKAADACRYLQTYGEWNRAAWLAKVRLNSEECADVLKRWVDHLCSPQV 1120
WD40-rpts
Query 1121 NQKSKALLVLLSLGCFVSVAETLHSMRYFDRAALFVEACLKYGAFEVSEDTEKLIAAIYADYARSLKSLGFRQGAVRFAS 1200
WD40-rpts
Query 1201 KAGAAGRDLLNELGSTKEELTES 1223
WD40-rpts
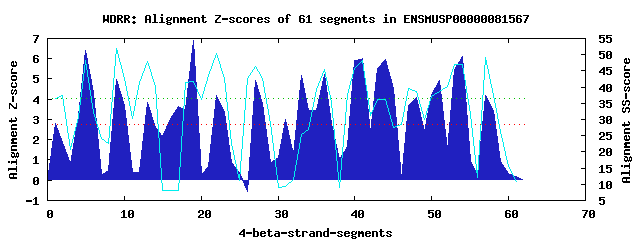 17 WD40-repeats found in QUERY=ENSMUSP00000081567 (Len=1223): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 8-47 2.80 230.47 0.04769 17.5% 36.48 WD 2: Seg 5 51-99 6.39 378.70 0.00049 30.0% 48.28 WD 3: Seg 9 103-145 4.96 319.49 0.00308 15.0% 51.86 WD 4: Seg 13 149-187 3.83 272.72 0.01308 17.5% 47.83 WD 5: Seg 17 190-228 3.62 264.36 0.01692 12.5% 8.17 WD 6: Seg 19 233-272 6.91 399.90 0.00025 12.5% 41.82 WD 7: Seg 22 273-316 4.18 287.13 0.00838 12.5% 50.40 WD 8: Seg 27 340-384 4.92 317.91 0.00323 12.5% 46.31 WD 9: Seg 31 387-431 3.02 239.30 0.03647 12.5% 9.20 WD10: Seg 33 456-500 5.16 327.66 0.00239 20.0% 25.32 WD11: Seg 36 504-550 5.22 330.24 0.00220 20.0% 45.57 WD12: Seg 41 560-595 6.01 362.96 0.00080 17.5% 48.27 WD13: Seg 44 599-637 5.94 359.89 0.00088 15.0% 36.18 WD14: Seg 48 653-691 4.09 283.69 0.00932 12.5% 38.74 WD15: Seg 51 694-735 4.91 317.62 0.00326 22.5% 38.79 WD16: Seg 54 738-777 6.12 367.58 0.00069 25.0% 46.94 WD17: Seg 57 781-821 4.18 287.38 0.00832 20.0% 49.09Multiple sequence alignment of 17 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 8-47 FKV----S--AR-----TLTG-----ALN-----AHNKAAVDW----GW----------QGLI--------AYGCHSLVVVID WD 2 51-99 -AQ----T--LQ-----VLEK-----HKA-----DIVKVRWAR----ENYHHNIGSPYCLRLA--------SADVTGKIIVWD WD 3 103-145 -GV----A--QC-----EIQE-----HVK-----PIQDVQWLWNQDASR----------DLLL--------AIHPPNYIVLWN WD 4 149-187 -GT----K--LW-----KKSY-----ADN-----ILSFSFDPF----DP----------SHLT--------LLTSEGIVFISD WD 5 190-228 -PS----K--PP-----SGPG-----KKV-----YISSPHSSP----AH----------NKLA--------AATGAKKALNKV WD 6 233-272 TQE----K--PS-----ADFV-----ALN-----DCLQLAYLP----SK----------RNHM--------LLLYPREILILD WD 7 273-316 LEVNQTVG--VI-----AIER-----TGV-----PFLQVIPCS----QR----------DGLF--------CLHENGCITLRV WD 8 340-384 TYD----L--RSQCDAIRVTK-----TVR-----PFSMVCCPV----NE----------NAAA--------LIVSDGRVMIWE WD 9 387-431 SAV----C--SR-----NARN-----SSGVSPLYSPVSFCGIP----GG----------VLQN--------KLPDLSLDNMIG WD10 456-500 TGL----L--SG-----LPSPQFAIRMCP-----PLTTKNIKM----YQ----------PLLA--------VGTSNGSVLVYH WD11 504-550 -GL----L--HK-----ELSV-----HSC-----EVKGIEWTS----LT----------SFLSFAASTPNNMGLVRNELQLVD WD12 560-595 RGD---------------RGN-----DES-----PIEMIKVSH----LK----------QYLA--------VVFKDKPLELWD WD13 599-637 -CT----L--LR-----EMSK-----SFP-----AITALEWSP----SH----------NLKS--------LRKKQLATREAM WD14 653-691 -SS----V--IS-----LLQE-----AES-----KAELSQNIS----AR----------EHFV--------FTDNDGQVYHLT WD15 694-735 GNS----VKDSA-----RIPP-----DGS-----MGSITCIAW----KG----------DTLV--------LGDMDGNLNFWD WD16 738-777 -AR----V--SR-----GIPT-----HRS-----WVRKIRFAP---GKG----------NQKL--------IAMYNDGAEVWD WD17 781-821 -VQ----M--VS-----SLRS-----GRN--VTFRILDVDWCT----SD----------K-VI--------LASDDGCIRVLE WD40 Template TGE----C--LR-----TLSG-----HTS-----GVTSVSFSP----DG----------KRLV--------SGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |