WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000078503 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000078503 (Len=468) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000078503, Len=468): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MEVHLPRLPLMKIFSYLDAYSLLQAAQVNKNWNELASSDVLWRKLCQKRWLFCDMVTLQLLGTETWKQFFVFRTWQEHAK 80
WD40-rpts (__
Query 81 TRAKPEDFTYKEIPAEYGFRANACYISGCGLTRNVQDRSVICMVSSMTKLSTWDIREGIMTWVSTEQPAYIKLLTTLPEM 160
WD40-rpts ________________________WD 1_________________________) (_________________WD 2_
Query 161 HIAVTVDTHSTIKLWDCHNREALATNCLFSSCKLLKAVFSKDDPIVLVGDISGNLYIFRIPDLHLISKVNVFPYGIDELH 240
WD40-rpts _______________) (_________________WD 3_________________) (_________________
Query 241 CSPQKKWVFLIMKHPRVLTKVVYMNSLLRTSEFSAPVSTILKFSLCDKAFWTPRREDRITQMSRGVPPRPTKFATFDMKL 320
WD40-rpts WD 4________________) (_________________WD 5________________)
Query 321 EEIGNKVTVQEYLVASFSLQDYEASPEWMGVSNKDVIVCSTGSSLLLFNIHGLRLQTFHYCPEEILRLWVDPLHVIVTCN 400
WD40-rpts (_________________WD 6________________) (_________________WD 7_______
Query 401 DGSMDVYAWEERSLLLRKCYRLQNRRNLPPESFIFKTLCDDVSIIGVMTNSPAPCFLMAYTLASALEN 468
WD40-rpts _________)
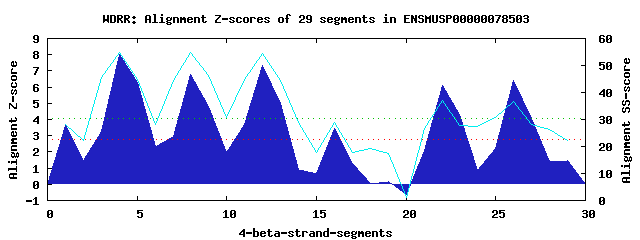 7 WD40-repeats found in QUERY=ENSMUSP00000078503 (Len=468): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 78-134 3.61 263.62 0.01731 17.5% 28.05 WD 2: Seg 4 138-176 8.04 446.72 5.9e-05 20.0% 54.87 WD 3: Seg 8 180-219 6.79 395.04 0.00030 22.5% 54.83 WD 4: Seg 12 223-261 7.36 418.68 0.00014 10.0% 54.62 WD 5: Seg 16 265-303 3.45 257.14 0.02113 20.0% 29.06 WD 6: Seg 22 331-369 6.07 365.28 0.00074 7.5% 36.85 WD 7: Seg 26 372-410 6.38 378.24 0.00050 12.5% 36.55Multiple sequence alignment of 7 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 78-134 HAKTRAKPEDFTYKEIPAEYGFRANACYIS-GCGLTRNVQDRSVICMVSSMTKLSTWD WD 2 138-176 -GIM-----------------TWVSTEQPA-YIKLLTTLPEMHIAVTVDTHSTIKLWD WD 3 180-219 -REA-----------------LATNCLFSSCKLLKAVFSKDDPIVLVGDISGNLYIFR WD 4 223-261 -LHL-----------------ISKVNVFPY-GIDELHCSPQKKWVFLIMKHPRVLTKV WD 5 265-303 -NSL-----------------LRTSEFSAP-VSTILKFSLCDKAFWTPRREDRITQMS WD 6 331-369 -EYL-----------------VASFSLQDY-EASPEWMGVSNKDVIVCSTGSSLLLFN WD 7 372-410 -GLR-----------------LQTFHYCPE-EILRLWVDPLHVIVTCNDGSMDVYAWE WD40 Template TGEC-----------------LRTLSGHTS-GVTSVSFSPDGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |