WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000072509 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000072509 (Len=1489) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000072509, Len=1489): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAGNSLVLPIVLWGRKAPTHCISSILLTDDGGTIVTGCHDGQICLWDVSVELEVNPRALLFGHTASITCLSKACASGDKR 80
WD40-rpts (_________________WD 1________________) (__________________WD 2____
Query 81 YTVSASANGEMCLWDVNDGRCIEFTKLACTHTGIQFYQFSVGNQQEGRLLCHGHYPEILVVDATSLEVLYSLVSKISPDW 160
WD40-rpts ______________) (_______________WD 3_______________) (______________
Query 161 ISSMSIIRSQRTQEDTVVALSVTGILKVWIVTSEMSGMQDTEPIFEEESKPIYCQNCQSISFCAFTQRSLLVVCSKYWRV 240
WD40-rpts ______WD 4___________________) (_____
Query 241 FDAGDYSLLCSGPSENGQTWTGGDFVSADKVIIWTENGQSYIYKLPASCLPASDSFRSDVGKAVENLIPPVQHSLLDQKD 320
WD40-rpts ____________WD 5________________) (___
Query 321 KELVICPPVTRFFYGCKEYLHKLLIQGDSSGRLNIWNIADIAEKQEADEGLKMTTCISLQEAFDKLKPCPAGIIDQLSVI 400
WD40-rpts _______________WD 6_________________) (_____________
Query 401 PNSNEPLKVTASVYIPAHGRLVCGREDGSIIIVPATQTAIVQLLQGEHMLRRGWPPHRTLRGHRNKVTCLLYPHQVSARY 480
WD40-rpts ________WD 7_____________________) (________________________WD 8_______
Query 481 DQRYLISGGVDFSVIIWDIFSGEMKHIFCVHGGEITQLLVPPENCSARVQHCICSVASDHSVGLLSLREKKCIMLASRHL 560
WD40-rpts _________________) (____________________WD 9___________________) (__________
Query 561 FPIQVIKWRPSDDYLVVGCTDGSVYVWQMDTGALDRCAMGITAVEILNACDEAVPAAVDSLSHPAVNLKQAMTRRSLAAL 640
WD40-rpts _______WD10________________) (_________________WD11________________)(_________
Query 641 KNMAHHKLQTLATNLLASEASDKGNLPKYSHNSLMVQAIKTNLTDPDIHVLFFDVEALIIQLLTEEASRPNTALISPENL 720
WD40-rpts ________WD12________________) (_________________WD13__
Query 721 QKASGSSDKGGSFLTGKRAAVLFQQVKETIKENIKEHLLDEEEDEEEARRQSREDSDPEYRASKSKPLTLLEYNLTMDTA 800
WD40-rpts ______________) (_______________
Query 801 KLFMSCLHAWGLNEVLDEVCLDRLGMLKPHCTVSFGLLSRGGHMSLMLPGYNQAAGKLLHAKAEVGRKLPAAEGVGKGTY 880
WD40-rpts ________________WD14_______________________________) (_______________________W
Query 881 TVSRAVTTQHLLSIISLANTLMSMTNATFIGDHMKKGPTRPPRPGTPDLSKARDSPPPSSNIVQGQIKQAAAPVVSARSD 960
WD40-rpts D15______________________)
Query 961 ADHSGSDSASPALPTCFLVNEGWSQLAAMHCVMLPDLLGLERFRPPLLEMLARRWQDRCLEVREAAQALLLAELRRIEQA 1040
WD40-rpts
Query 1041 GRKETIDTWAPYLPQYMDHVISPGVTAEAMQTMAAAPDASGPEAKVQEEEHDLVDDDITAGCLSSVPQMKKISTSYEERR 1120
WD40-rpts
Query 1121 KQATAIVLLGVIGAEFGAEIEPPKLLTRPRSSSQIPEGFGLTSGGSNYSLARHTCKALTYLLLQPPSPKLPPHSTIRRTA 1200
WD40-rpts
Query 1201 TDLIGRGFTVWEPYMDVSAVLMGLLELCADAEKQLANITMGLPLSPAADSARSARHALSLIATARPPAFITTIAKEVHRH 1280
WD40-rpts
Query 1281 TALAANTQSQQSIHTTTLARAKGEILRVIEILIEKMPTDVVDLLVEVMDIIMYCLEGSLVKKKGLQECFPAICRFYMVSY 1360
WD40-rpts (_________________W
Query 1361 YERSHRIAVGARHGSVALYDIRTGKCQTIHGHKGPITAVSFAPDGRYLATYSNTDSHISFWQMNTSLLGSIGMLNSAPQL 1440
WD40-rpts D16________________) (_________________WD17_________________)(_________________
Query 1441 RCIKTYQVPPVQPASPGSHNALKLARLIWTSNRNVILMAHDGKEHRFMV 1489
WD40-rpts __WD18___________________)
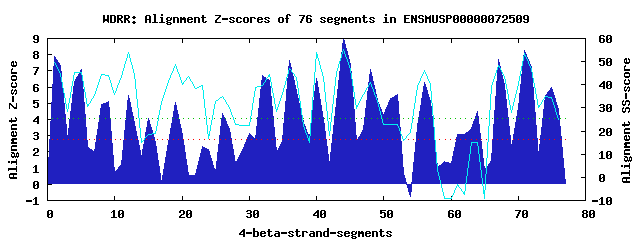 18 WD40-repeats found in QUERY=ENSMUSP00000072509 (Len=1489): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 9-47 7.90 441.08 7.1e-05 30.0% 49.80 WD 2: Seg 5 54-95 7.08 407.17 0.00020 35.0% 45.18 WD 3: Seg 9 107-142 5.13 326.55 0.00247 25.0% 43.92 WD 4: Seg 12 146-190 5.48 341.23 0.00157 30.0% 53.78 WD 5: Seg 19 235-273 5.07 323.97 0.00268 15.0% 48.74 WD 6: Seg 26 317-357 4.38 295.48 0.00647 12.5% 35.12 WD 7: Seg 32 387-434 6.74 392.87 0.00032 30.0% 39.70 WD 8: Seg 36 445-498 7.65 430.65 9.8e-05 40.0% 47.24 WD 9: Seg 40 502-546 6.51 383.76 0.00042 17.5% 53.86 WD10: Seg 44 550-588 8.97 485.11 1.8e-05 27.5% 55.23 WD11: Seg 48 592-630 7.11 408.17 0.00020 12.5% 41.10 WD12: Seg 52 631-669 5.55 343.96 0.00144 12.5% 22.96 WD13: Seg 56 697-735 6.30 374.76 0.00055 20.0% 45.99 WD14: Seg 61 785-852 3.08 241.81 0.03378 10.0% -2.92 WD15: Seg 64 856-906 4.47 299.17 0.00577 12.5% 14.89 WD16: Seg 67 1342-1380 7.71 433.28 9.0e-05 17.5% 48.32 WD17: Seg 71 1383-1422 8.28 456.55 4.4e-05 37.5% 53.60 WD18: Seg 75 1423-1466 5.96 360.82 0.00085 7.5% 34.38Multiple sequence alignment of 18 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 9-47 -PI----VL--------WGR--------KA-PT-----------------------------HC--ISSIL---LT------DDG------GT----IVTGCH------------DGQICLWD WD 2 54-95 -VN----PR--------ALL--------FG-HT-----------------------------AS--ITCLSKACAS------GDK------RY----TVSASA------------NGEMCLWD WD 3 107-142 -------LA--------CTH--------TG-IQ-----------------------------FY--QFSVG---NQ------QEG------RL----LCHGHY------------P-EILVVD WD 4 146-190 -LE----VL--------YSL--------VS-KI-----------------------------SPDWISSMS---II------RSQ------RTQEDTVVALSV------------TGILKVWI WD 5 235-273 -SK----YW--------RVF--------DA-GD-----------------------------YS--LLCSG---PS------ENG------QT----WTGGDF------------VSADKVII WD 6 317-357 DQK----DK--------ELV--------ICPPV-----------------------------TR--FFYGC---KE------YLH------KL----LIQGDS------------SGRLNIWN WD 7 387-434 KPC----PA--------GIIDQLSVIPNSN-EP-----------------------------LK--VTASV---YI------PAH------GR----LVCGRE------------DGSIIIVP WD 8 445-498 QGE----HMLRRGWPPHRTL--------RG-HR-----------------------------NK--VTCLL---YPHQVSARYDQ------RY----LISGGV------------DFSVIIWD WD 9 502-546 -GE----MK--------HIF--------CV-HG-----------------------------GE--ITQLL---VP------PENCSARVQHC----ICSVAS------------DHSVGLLS WD10 550-588 -KK----CI--------MLA--------SR-HL-----------------------------FP--IQVIK---WR------PSD------DY----LVVGCT------------DGSVYVWQ WD11 592-630 -GA----LD--------RCA--------MG-IT-----------------------------AV--EILNA---CD------EAV------PA----AVDSLS------------HPAVNLKQ WD12 631-669 -AM----TR--------RSL--------AA-LK-----------------------------NM--AHHKL---QT------LAT------NL----LASEAS------------DKGNLPKY WD13 697-735 -AL----II--------QLL--------TE-EA-----------------------------SR--PNTAL---IS------PEN------LQ----KASGSS------------DKGGSFLT WD14 785-852 -SK----PL--------TLL--------EY-NLTMDTAKLFMSCLHAWGLNEVLDEVCLDRLGM--LKPHC---TV------SFG------LL----SRGGHM------------SLMLPGYN WD15 856-906 -GK----LL--------HAK--------AE-VG-----------------------------RK--LPAAE---GV------GKG------TY----TVSRAVTTQHLLSIISLANTLMSMTN WD16 1342-1380 -KK----GL--------QEC--------FP-AI-----------------------------CR--FYMVS---YY------ERS------HR----IAVGAR------------HGSVALYD WD17 1383-1422 -TG----KC--------QTI--------HG-HK-----------------------------GP--ITAVS---FA------PDG------RY----LATYSN-----------TDSHISFWQ WD18 1423-1466 MNTSLLGSI--------GML--------NS-AP-----------------------------QL--RCIKT---YQ------VPP------VQ----PASPGS------------HNALKLAR WD40 Template TGE----CL--------RTL--------SG-HT-----------------------------SG--VTSVS---FS------PDG------KR----LVSGSE------------DGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |