WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000051080 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000051080 (Len=1958) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000051080, Len=1958): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MADRTAPRCQLRLEWVYGYRGHQCRNNLYYTAGKEVVYFVAGVGVVYNTREHSQKFFLGHNDDIISLALHPDKTLIATGQ 80
WD40-rpts (__________________WD 1_______
Query 81 VGKEPYICIWNSYNVHTVSILKDVHTHGVACLAFDSDGQHLASVGLDAKNTVCIWDWRKGKLLASATGHSDRIFDISWDP 160
WD40-rpts __________)(____________________WD 2___________________) (_________________WD
Query 161 YQPNRMVSCGVKHIKFWTLCGNALTAKRGIFGKTGDLQTILCLACAKEDITYSGALNGDIYVWKGLTLVRTIQGAHSAGI 240
WD40-rpts 3________________) (________________WD 4________________)(_______________
Query 241 FSLYACEEGFATGGRDGCIRLWDTDFKPITKIDLRETEQGYKGLSIRSVCWKADRLLAGTQDSEIFEVIVRERDKPMLIL 320
WD40-rpts __WD 5________________) (__________________WD 6_________________) (______
Query 321 QGHCEGELWALALHPKKPLAVTGSDDRSVRLWSLADHALIARCNMEEAVRSVSFSPDGSQLALGMKDGSFIVLRVRDMTE 400
WD40-rpts ___________WD 7_________________)(__________________WD 8_________________) (__
Query 401 VVHIKDRKEVIHEMKFSPDGSYLAVGSNDGPVDVYAVAQRYKKIGECNKSLSFITHIDWSLDSKYLQTNDGAGERLFYKM 480
WD40-rpts _______________WD 9________________) (_________________WD10________________)
Query 481 PSGKPLTSKEEIKGIPWASWTCVRGPEVSGIWPKYTEVIDINSVDANYNSSVLVSGDDFGLVKLFKFPCLKKGAKFRKYV 560
WD40-rpts (___________________WD11__________________) (_______
Query 561 GHSAHVTNVRWSHDFQWVLSTGGADHSVFQWRFIPEAVSNGVLETTPQEGGADSYSEESDSDFSDVPELDSDIEQETQIN 640
WD40-rpts __________WD12_________________) (_________________________WD13_________________
Query 641 YDRQVYKEDLPQLKQQSKEKNHAVPFLKREKAPEDSLKLQFIHGYRGYDCRNNLFYTQAGEVVYHIAAVAVVYNRQQHAQ 720
WD40-rpts ________) (___
Query 721 RLYLGHDDDILSLTIHPVKDYVATGQVGRDAAVHVWDTQTLKCLSLLKGHHQRGVCALDFSADGKCLVSVGLDDFHSVVF 800
WD40-rpts _______________WD14_________________) (__________________WD15_________________
Query 801 WDWKKGEKIATTRGHKDKIFVVKCNPQHADKLVTVGIKHIKFWQQAGGGFTSKRGSFGSAGKLETMMCVSYGRMEDLVFS 880
WD40-rpts _) (_________________WD16________________) (________________WD17_____
Query 881 GAATGDIFIWKDVLLLKTVKAHDGPVFAMYALDKGFVTGGKDGIVELWDDMFERCLKTYAIKRTALSTSSKGLLLEDNPS 960
WD40-rpts __________)(________________WD18________________)(_________________WD19_________
Query 961 IRAITLGHGHILVGTKNGEILEIDKSGPMTLLVQGHMEGEVWGLAAHPLLPICATVSDDKTLRIWELSSQHRMLAVRKLK 1040
WD40-rpts _______) (_________________WD20_________________) (__________
Query 1041 KGGRCCAFSPDGKALAVGLNDGSFLVVNADTVEDMLSFHHRKEMISDIKFSKDTGKYLAVASHDNFVDIYNVLTSKRVGI 1120
WD40-rpts _______WD21________________) (_________________WD22_________________) (_____
Query 1121 CKGASSYITHIDWDSRGKLLQVNSGAKEQLFFEAPRGRKHTIRPSEAEKIEWDTWTCVLGPTCEGIWPAHSDVTDVNAAN 1200
WD40-rpts ____________WD23________________) (_____________________WD24_
Query 1201 LTKDGSLLATGDDFGFVKLFSYPVKGQHARFKKYVGHSAHVTNVRWLHNDSVLLTVGGADTALMIWTREFVGTQESKLVD 1280
WD40-rpts ____________________) (_________________WD25_________________) (______
Query 1281 SEESDTDAEEDGGYDSDVAREKAIDYTTKIYAVSIREMEGTKPHQQLKEVSMEERPPVSRAAPQPEKLQKNNITKKKKLV 1360
WD40-rpts ___________WD26________________) (__________________WD27_________________
Query 1361 EELALDHVFGYRGFDCRNNLHYLNDGADIIFHTAAAGIVQNLSTGSQSFYLEHTDDILCLTVNQHPKYRNVVATSQIGTT 1440
WD40-rpts ) (________________WD28_______________)(____________________WD29______________
Query 1441 PSIHIWDAMTKHTLSMLRCFHTKGVNYINFSATGKLLVSVGVDPEHTITVWRWQEGTKVASRGGHLERIFVVEFRPDSDT 1520
WD40-rpts ______) (__________________WD30_________________) (_________________WD31___
Query 1521 QFVSVGVKHMKFWTLAGSALLYKKGVIGSMEAAKMQTMLSVAFGANNLTFTGAINGDVYVWKEHFLIRLVAKAHTGPVFT 1600
WD40-rpts _____________) (_______________WD32_______________)(_________________
Query 1601 MYTTLRDGLIVTGGKERPTKEGGAVKLWDQEMKRCRAFQLETGQLVECVRSVCRGKGKILVGTKDGEIIEVGEKSAASNI 1680
WD40-rpts ____WD33____________________)(___________________WD34___________________) (____
Query 1681 LIDGHMEGEIWGLATHPSKDMFISASNDGTARIWDLADKKLLNKVNLGHAARCAAYSPDGEMVAIGMKNGEFVILLVNTL 1760
WD40-rpts _____________WD35_________________)(__________________WD36_________________) (
Query 1761 KVWGKKRDRKSAIQDIRISPDNRFLAVGSSEQTVDFYDLTQGTSLNRIGYCKDIPSFVIQMDFSADSKYIQVSTGAYKRQ 1840
WD40-rpts _________________WD37________________) (_______________WD38_____________
Query 1841 VHEVPLGKQVTEAMVVEKITWASWTSVLGDEVIGIWPRNADKADVNCACVTHAGLNIVTGDDFGLLKLFDFPCTEKFAKH 1920
WD40-rpts __) (_____________________WD39_____________________) (___
Query 1921 KRYFGHSAHVTNIRFSSDDKYVVSTGGDDCSVFVWRCL 1958
WD40-rpts ______________WD40_________________)
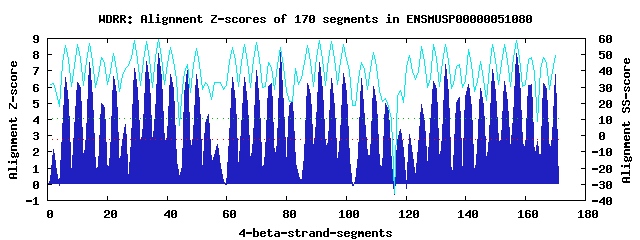 40 WD40-repeats found in QUERY=ENSMUSP00000051080 (Len=1958): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 6 51-91 6.61 387.59 0.00037 20.0% 55.90 WD 2: Seg 10 92-136 6.29 374.67 0.00056 32.5% 56.54 WD 3: Seg 14 140-178 7.49 424.09 0.00012 22.5% 56.91 WD 4: Seg 18 187-224 4.97 319.90 0.00304 17.5% 48.39 WD 5: Seg 22 225-263 6.66 389.61 0.00035 30.0% 50.86 WD 6: Seg 26 270-310 3.67 266.34 0.01592 25.0% 41.75 WD 7: Seg 29 314-353 7.15 410.04 0.00019 25.0% 58.70 WD 8: Seg 33 354-394 7.61 428.97 0.00010 37.5% 57.98 WD 9: Seg 37 398-436 8.05 447.10 5.9e-05 27.5% 59.10 WD10: Seg 41 441-479 6.79 394.96 0.00030 17.5% 54.65 WD11: Seg 47 504-546 7.00 403.85 0.00022 22.5% 44.01 WD12: Seg 50 553-592 6.76 393.77 0.00031 32.5% 53.91 WD13: Seg 54 594-649 4.31 292.59 0.00708 10.0% 30.27 WD14: Seg 62 717-757 6.59 387.01 0.00038 25.0% 54.53 WD15: Seg 66 761-802 6.58 386.45 0.00039 45.0% 56.20 WD16: Seg 70 806-844 7.00 403.62 0.00023 25.0% 57.45 WD17: Seg 75 855-891 6.24 372.44 0.00060 20.0% 41.97 WD18: Seg 78 892-929 8.15 451.37 5.1e-05 27.5% 54.24 WD19: Seg 82 930-968 5.16 327.69 0.00239 20.0% 21.04 WD20: Seg 87 987-1026 6.22 371.46 0.00061 25.0% 55.44 WD21: Seg 91 1030-1068 7.49 424.09 0.00012 30.0% 58.81 WD22: Seg 95 1072-1111 6.55 385.04 0.00040 22.5% 57.95 WD23: Seg 99 1115-1153 6.82 396.44 0.00028 17.5% 56.00 WD24: Seg105 1174-1221 6.55 385.09 0.00040 22.5% 45.40 WD25: Seg109 1228-1267 6.05 364.62 0.00076 25.0% 51.67 WD26: Seg113 1274-1312 4.97 320.03 0.00303 15.0% 22.86 WD27: Seg118 1321-1361 3.36 253.56 0.02358 10.0% 28.14 WD28: Seg121 1365-1401 3.06 240.90 0.03474 12.5% 49.33 WD29: Seg125 1402-1447 4.90 317.12 0.00331 25.0% 55.19 WD30: Seg129 1452-1492 6.36 377.19 0.00051 40.0% 56.36 WD31: Seg133 1496-1534 7.42 421.34 0.00013 22.5% 56.45 WD32: Seg138 1547-1582 5.35 335.73 0.00186 20.0% 43.85 WD33: Seg141 1583-1629 6.16 369.11 0.00066 25.0% 53.00 WD34: Seg145 1630-1673 5.89 358.09 0.00093 17.5% 45.98 WD35: Seg149 1676-1715 7.07 406.59 0.00021 27.5% 56.17 WD36: Seg153 1716-1756 6.33 376.15 0.00053 17.5% 56.28 WD37: Seg157 1760-1798 8.08 448.49 5.6e-05 22.5% 58.55 WD38: Seg162 1808-1843 6.19 370.50 0.00063 22.5% 48.11 WD39: Seg166 1863-1910 6.20 370.61 0.00063 17.5% 48.51 WD40: Seg170 1917-1956 6.75 393.56 0.00031 35.0% 49.49Multiple sequence alignment of 40 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 51-91 EH----S------------QK----------------FF---L-GHND-D-IISLAL-H--PD-KT-LIAT-GQVGKE------P--Y---ICIWN WD 2 92-136 SY----NVHTV--------SI----------------LK---D-VHTH-G-VACLAF-D--SD-GQ-HLAS-VG--LD------AKNT---VCIWD WD 3 140-178 -G----K---L--------LA----------------SA---T-GHSD-R-IFDISW-D--PY-QP-NRMV-SC--GV------K--H---IKFWT WD 4 187-224 -K----R---G--------IF----------------GK---T-GDLQ-T-ILCLAC-A--KE-D--ITYS-GA--LN------G--D---IYVWK WD 5 225-263 GL----T---L--------VR----------------TI--QG-AHSA-G-IFSLYA-C--EE-G---FAT-GG--RD------G--C---IRLWD WD 6 270-310 TK----I---D--------LR---------------ETE---Q-GYKGLS-IRSVCW-K--AD-R--LLAG-TQ--DS------E--I---FEVIV WD 7 314-353 -D----K---P--------ML----------------IL---Q-GHCE-GELWALAL-H--PK-KP-LAVT-GS--DD------R--S---VRLWS WD 8 354-394 LA----D--HA--------LI----------------AR---C-NMEE-A-VRSVSF-S--PD-GS-QLAL-GM--KD------G--S---FIVLR WD 9 398-436 -M----T---E--------VV----------------HI---K-DRKE-V-IHEMKF-S--PD-GS-YLAV-GS--ND------G--P---VDVYA WD10 441-479 -Y----K---K--------IG----------------EC---N-KSLS-F-ITHIDW-S--LD-SK-YLQT-ND--GA------G--E---RLFYK WD11 504-546 RG----P---E--------VS----------------GIWPKY-TEVI-D-INSVDA-N--YN-SS-VLVS-GD--DF------G--L---VKLFK WD12 553-592 -G----A---K--------FR----------------KY---V-GHSA-H-VTNVRW-S--HD-FQWVLST-GG--AD------H--S---VFQWR WD13 594-649 IP----E---A--------VSNGVLETTPQEGGADSYSE---E-SDSD-F-SDVPEL-D--SD-IE-QETQ-IN--YD------R--Q---VYKED WD14 717-757 QH--------A--------QR----------------LY---L-GHDD-D-ILSLTI-H--PV-KD-YVAT-GQVGRD------A--A---VHVWD WD15 761-802 -L----K---C--------LS----------------LL---K-GHHQRG-VCALDF-S--AD-GK-CLVS-VG--LD------DFHS---VVFWD WD16 806-844 -G----E---K--------IA----------------TT---R-GHKD-K-IFVVKC-N--PQ-HA-DKLV-TV--GI------K--H---IKFWQ WD17 855-891 ----------G--------SF----------------GS---A-GKLE-T-MMCVSY-G--RM-ED-LVFS-GA--AT------G--D---IFIWK WD18 892-929 DV----L---L--------LK----------------TV---K-AHDG-P-VFAMYA-L--DK-G---FVT-GG--KD------G--I---VELWD WD19 930-968 -D----M---F--------ER----------------CL---K-TYAI-K-RTALST-S--SK-GL-LLED-NP--SI------R--A---ITLGH WD20 987-1026 -G----P---M--------TL----------------LV---Q-GHME-GEVWGLAA-H--PL-LP-ICAT-VS--DD------K--T---LRIWE WD21 1030-1068 -Q----H---R--------ML----------------AV---R-KLKK-G-GRCCAF-S--PD-GK-ALAV-GL--ND------G--S---FLVVN WD22 1072-1111 -V----E---D--------ML----------------SF---H-HRKE-M-ISDIKF-S--KDTGK-YLAV-AS--HD------N--F---VDIYN WD23 1115-1153 -S----K---R--------VG----------------IC---K-GASS-Y-ITHIDW-D--SR-GK-LLQV-NS--GA------K--E---QLFFE WD24 1174-1221 TW----T---CVLGPTCEGIW----------------PA---H-SDVT-D-VNAANL-T--KD-GS-LLAT-GD--DF------G--F---VKLFS WD25 1228-1267 -H----A---R--------FK----------------KY---V-GHSA-H-VTNVRWLH--ND-SV-LLTV-GG--AD------T--A---LMIWT WD26 1274-1312 -Q----E---S--------KL----------------VD---S-EESD-T-DAEEDG-G--YD-SD-VARE-KA--ID------Y--T---TKIYA WD27 1321-1361 TK----P---H--------QQ----------------LK---EVSMEE-R-PPVSRA-A--PQ-PE-KLQK-NN--IT------K--K---KKLVE WD28 1365-1401 -L----D---H--------VF----------------GY---R-GFDC---RNNLHY-L--ND-GA-DIIF-HT--AA------A--G---I-VQN WD29 1402-1447 LS----T--GS--------QS----------------FY---L-EHTD-D-ILCLTV-NQHPK-YR-NVVA-TS--QI------G--TTPSIHIWD WD30 1452-1492 -H----T---L--------SM----------------LR---C-FHTK-G-VNYINF-S--AT-GK-LLVS-VG--VD----PEH--T---ITVWR WD31 1496-1534 -G----T---K--------VA----------------SR---G-GHLE-R-IFVVEF-R--PD-SD-TQFV-SV--GV------K--H---MKFWT WD32 1547-1582 IG----S------------------------------ME---A-AKMQ-T-MLSVAF-G--AN-N--LTFT-GA--IN------G--D---VYVWK WD33 1583-1629 EH----F---L--------IR---------------LVA---K-AHTG-P-VFTMYT-T--LR-DG-LIVT-GG--KERPTKEGG--A---VKLWD WD34 1630-1673 QEMKRCR---A--------FQ----------------LE---T-GQLV-E-CVRSVC-R--GK-GK-ILVG-TK--DG------E--I---IEVGE WD35 1676-1715 -A----A---S--------NI----------------LI---D-GHME-GEIWGLAT-H--PS-KD-MFIS-AS--ND------G--T---ARIWD WD36 1716-1756 LA----D--KK--------LL----------------NK---V-NLGH-A-ARCAAY-S--PD-GE-MVAI-GM--KN------G--E---FVILL WD37 1760-1798 -L----K---V--------WG----------------KK---R-DRKS-A-IQDIRI-S--PD-NR-FLAV-GS--SE------Q--T---VDFYD WD38 1808-1843 IG----Y---C-------------------------------K-DIPS-F-VIQMDF-S--AD-SK-YIQV-ST--GA------Y--K---RQVHE WD39 1863-1910 SW----T---SVLGDEVIGIW----------------PR---N-ADKA-D-VNCACV-T--HA-GL-NIVT-GD--DF------G--L---LKLFD WD40 1917-1956 -F----A---K--------HK----------------RY---F-GHSA-H-VTNIRF-S--SD-DK-YVVSTGG--DD------C--S---VFVWR WD40 Template TG----E---C--------LR----------------TL---S-GHTS-G-VTSVSF-S--PD-GK-RLVS-GS--ED------G--T---IKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |