WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000047825 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000047825 (Len=532) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000047825, Len=532): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MKTSEERLLAPDSLPPDLAPAPVPQGSPAEKNTDFEPVPPPCGGDDQPQLATDPVASLVVSQELQQGDSVPLEVEFNTSS 80
WD40-rpts (____________________WD 1____________________) (______________WD 2____
Query 81 ELSPGIEEQDVSEHASLPGEETNLPELESGEATEGVSEERAEVDEGDTFWTYSFSQVPRYLSGSWSEFSTRSENFLKGCK 160
WD40-rpts _________)(__________________WD 3_________________) (_________________W
Query 161 WAPDGSCILTNSADNVLRIYNLPPELYSEQEQVDYAEMVPVLRMVEGDTIYDYCWYSLMSSTQPDTSYVASSSRENPIHI 240
WD40-rpts D 4_________________) (________________WD 5______________
Query 241 WDAFTGELRASFRAYNHLDELTAAHSLCFSPDGSQLFCGFNRTVRVFSTSRPGRDCEVRATFAKKQGQSGIISCIAFSPS 320
WD40-rpts _) (___________________WD 6__________________) (_________________WD 7
Query 321 QPLYACGSYGRTIGLYAWDDGSPLALLGGHQGGITHLCFHPDGNLFFSGARKDAELLCWDLRQPGHLLWSLSREVTTNQR 400
WD40-rpts ________________) (_________________WD 8_________________) (_______________
Query 401 IYFDLDPSGQFLVSGNTSGVVSVWDISGALSDDSKLEPVVTFLPQKDCTNGVSLHPTLPLLATASGQRVFPEPTNSGDEG 480
WD40-rpts ___WD 9_________________)(______________________WD10_____________________)
Query 481 ELELELPLLSLCHAHPECQLQLWWCGGGPDPSSPVDDQDEKGQRRTEAVGMS 532
WD40-rpts
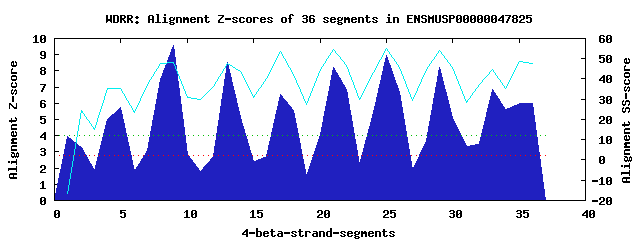 10 WD40-repeats found in QUERY=ENSMUSP00000047825 (Len=532): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 7-52 3.95 277.98 0.01112 10.0% -16.30 WD 2: Seg 2 58-90 3.26 249.26 0.02691 17.5% 24.57 WD 3: Seg 5 91-131 5.72 351.08 0.00115 12.5% 35.38 WD 4: Seg 9 142-181 9.54 508.83 8.6e-06 15.0% 48.12 WD 5: Seg 13 206-242 8.49 465.37 3.3e-05 25.0% 47.79 WD 6: Seg 17 246-288 6.57 385.90 0.00039 30.0% 53.42 WD 7: Seg 21 299-337 8.20 453.32 4.8e-05 20.0% 54.59 WD 8: Seg 25 341-380 8.97 485.35 1.8e-05 40.0% 55.14 WD 9: Seg 29 385-425 8.19 452.90 4.9e-05 37.5% 54.06 WD10: Seg 33 426-474 6.84 397.32 0.00027 17.5% 44.68Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 7-52 RL---------LAPDSLPPDL-----APAP--VPQG--SPAEKNTDFEPVPPPCG-GDDQPQLAT WD 2 58-90 -L---------VVSQELQQGD-----SVPL--EVEF--NT------------SSE-LSPGIEEQD WD 3 91-131 -V---------SEHASLPGEE-----TNLPELESGE--AT------EGVSEERAE-VDEGDTFWT WD 4 142-181 SG---------SWSEFSTRSE-----NFLK--GCKW--AP------DGSCILTNS-ADNVLRIYN WD 5 206-242 EG---------D---TIYDYC-----WYSL--MSST--QP------DTSYVASSS-RENPIHIWD WD 6 246-288 -G---------ELRASFRAYNHLDELTAAH--SLCF--SP------DGSQLFCGF--NRTVRVFS WD 7 299-337 -R---------ATFAKKQGQS-----GIIS--CIAF--SP------SQPLYACGS-YGRTIGLYA WD 8 341-380 -G---------SPLALLGGHQ-----GGIT--HLCF--HP------DGNLFFSGARKDAELLCWD WD 9 385-425 -G---------HLLWSLSREV-----TTNQ--RIYFDLDP------SGQFLVSGN-TSGVVSVWD WD10 426-474 ISGALSDDSKLEPVVTFLPQK-----DCTN--GVSL--HP------TLPLLATAS-GQRVFPEPT WD40 Template TG---------ECLRTLSGHT-----SGVT--SVSF--SP------DGKRLVSGS-EDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |