WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000033506 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000033506 (Len=485) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000033506, Len=485): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAAVWQQVLAVDARYNAYRTPTFPQFRTQYIRRRSQLLRENAKAGHPPALRRQYLRLRGQLLGQRYGPLSEPGSARAYSN 80
WD40-rpts (____________WD 1_______
Query 81 SIVRSSRTTLDRMEDFEDDPRALGARGHRRSVSRGSYQLQAQMNRAVYEDRPPGSVVPTSVAEASRAMAGDTSLSENYAF 160
WD40-rpts _____) (_______________WD 2______________)
Query 161 AGMYHVFDQHVDEAVPRVRFANDDRHRLACCSLDGSISLCQLVPAPPTVLHVLRGHTRGVSDFAWSLSNDILVSTSLDAT 240
WD40-rpts (_________________WD 3_________________) (_________________WD 4____________
Query 241 MRIWASEDGRCIREIPDPDGAELLCCTFQPVNNNLTVVGNAKHNVHVMNISTGKKVKGGSSKLTGRVLALSFDAPGRLLW 320
WD40-rpts ____) (__________________WD 5_________________) (__________________WD 6_____
Query 321 AGDDRGSVFSFLFDMATGKLTKAKRLVVHEGSPVTSISARSWVSREARDPSLLINACLNKLLLYRVVDNEGALQLKRSFP 400
WD40-rpts _____________) (__________________WD 7__________________)(___________
Query 401 IEQSSHPVRSIFCPLMSFRQGACVVTGSEDMCVHFFDVERAAKAAVNKLQGHSAPVLDVSFNCDESLLASSDASGMVIVW 480
WD40-rpts ___________WD 8_____________________) (_________________WD 9________________
Query 481 RREQK 485
WD40-rpts )
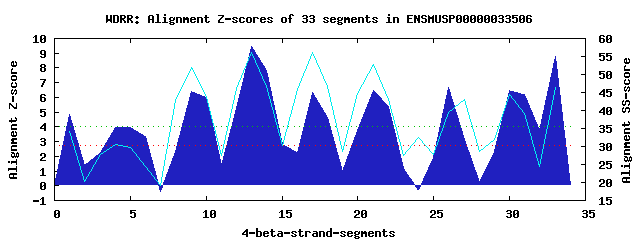 9 WD40-repeats found in QUERY=ENSMUSP00000033506 (Len=485): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 57-86 4.82 313.82 0.00367 5.0% 34.12 WD 2: Seg 5 103-137 3.96 278.22 0.01103 15.0% 29.61 WD 3: Seg 9 162-201 6.41 379.44 0.00048 27.5% 51.85 WD 4: Seg 13 207-245 9.47 505.93 9.4e-06 37.5% 56.25 WD 5: Seg 17 249-289 6.32 375.53 0.00054 22.5% 56.11 WD 6: Seg 21 293-334 6.45 381.06 0.00046 27.5% 52.90 WD 7: Seg 26 347-388 6.66 389.85 0.00035 10.0% 39.34 WD 8: Seg 30 389-437 6.49 382.57 0.00043 25.0% 44.51 WD 9: Seg 33 443-481 8.79 477.58 2.3e-05 32.5% 46.37Multiple sequence alignment of 9 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 57-86 ---------------L-RGQ-LLGQ----RYGPLSE-PG-SARAYSNSI--VRSS WD 2 103-137 -----L-----GARGH-RRS-VSRG----SYQLQAQ-MN-RAVYEDRPP--GSVV WD 3 162-201 -GMYHV-----FDQHV-DEA-VPRV----RFANDDRHRL-ACCSLDGSI--SLCQ WD 4 207-245 -PTVLH-----VLRGH-TRG-VSDF----AWSLSND-IL-VSTSLDATM--RIWA WD 5 249-289 -GRCIR-----EIPDP-DGAELLCC----TFQPVNN-NLTVVGNAKHNV--HVMN WD 6 293-334 -GKKVK-----GGSSKLTGR-VLAL----SFDAPGR-LL-WAGDDRGSVFSFLFD WD 7 347-388 VVHEGS-----PVTSI-SAR-SWVS----REARDPS-LL-INACLNKLLLYRVVD WD 8 389-437 NEGALQLKRSFPIEQS-SHP-VRSIFCPLMSFRQGA-CV-VTGSEDMCV--HFFD WD 9 443-481 -KAAVN-----KLQGH-SAP-VLDV----SFNCDES-LL-ASSDASGMV--IVWR WD40 Template TGECLR-----TLSGH-TSG-VTSV----SFSPDGK-RL-VSGSEDGTI--KVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |