WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSMUSP00000030895 [Mus musculus] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSMUSP00000030895 (Len=462) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSMUSP00000030895, Len=462): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MNFSESFKLSGLLCRFSPDGKYLASCVQYRLVIRDVTTLQILQLYTCLDQIQHIEWSADSLFILCAMYRRGLVQVWSLEQ 80
WD40-rpts (_______________WD 1______________) (_________________WD 2_________________)
Query 81 PEWHCKIDEGSAGLVASCWSPDGRHILNTTEFHLRITVWSLCTKSVSYIKYPKACQQGLTFTRDGRYLALAERRDCRDYV 160
WD40-rpts (_________________WD 3_________________) (__________________WD 4_______________
Query 161 SIFVCSDWQLLRHFDTDTQDLTGIEWAPNGCVLAAWDTCLEYKVLLYSLDGRLLSAYCAYEWSLGIKSVAWSPSSQFLAI 240
WD40-rpts ___) (__________________WD 5_________________) (__________________WD 6_______
Query 241 GSYDGKVRLLNHVTWKMITEFGHPATINNPKTVVYKEAEKSPLLGLGHLSFPPPRAMAGALSTSESKYEIASGPVSLQTL 320
WD40-rpts __________) (__________________WD 7_________________) (_
Query 321 KPVADRANPRMGVGMLAFSSDSYFLASRNDNVPNAVWIWDIQKLKLFVVLEHMSPVRSFQWDPQQPRLAICTGGSKVYLW 400
WD40-rpts _________________WD 8__________________)(__________________WD 9_________________
Query 401 SPAGCVSVQVPGEGDFPVLGLCWHLSGDSLALLSKDHFCLCFLETKERVGTAYEQRDGMPRT 462
WD40-rpts ) (_________________WD10________________)
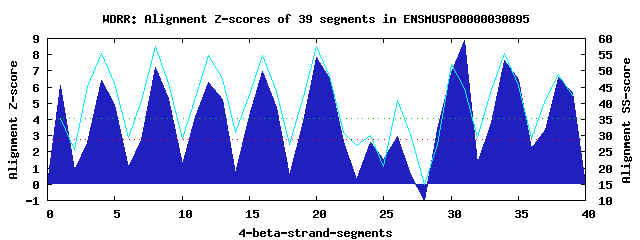 10 WD40-repeats found in QUERY=ENSMUSP00000030895 (Len=462): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-35 6.07 365.47 0.00074 22.5% 35.43 WD 2: Seg 4 38-77 6.43 380.47 0.00046 15.0% 55.24 WD 3: Seg 8 81-120 7.24 413.57 0.00017 25.0% 57.46 WD 4: Seg 12 123-164 6.29 374.44 0.00056 10.0% 54.75 WD 5: Seg 16 168-208 6.99 403.57 0.00023 17.5% 54.84 WD 6: Seg 20 211-251 7.80 436.94 8.0e-05 30.0% 57.53 WD 7: Seg 26 269-309 2.94 235.91 0.04044 15.0% 40.97 WD 8: Seg 31 319-360 8.85 480.09 2.1e-05 25.0% 44.68 WD 9: Seg 34 361-401 7.67 431.52 9.5e-05 22.5% 55.39 WD10: Seg 38 405-443 6.61 387.73 0.00037 12.5% 48.98Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-35 -----MNFSESF--KL---SGLLCRFSPDGKY-LASCV-Q--Y--RL---V-IRD WD 2 38-77 -TLQ-ILQLYTC--LD---QIQHIEWSADSLF-ILCAMYR--R--GL---VQVWS WD 3 81-120 -PEW-HCKIDEG--SA---GLVASCWSPDGRHILNTTE-F--H--LR---ITVWS WD 4 123-164 -TKS-VSYIKYP--KA---CQQGLTFTRDGRY-LALAE-R--R--DCRDYVSIFV WD 5 168-208 -WQL-LRHFDTD--TQ---DLTGIEWAPNGCV-LAAWD-TCLE--YK---VLLYS WD 6 211-251 -GRL-LSAYCAYEWSL---GIKSVAWSPSSQF-LAIGS-Y--D--GK---VRLLN WD 7 269-309 NPKT-VVYKEAE--KSPLLGLGHLSFPPPRA---MAGA-L--S--TS---ESKYE WD 8 319-360 TLKP-VADRANP--RM---GVGMLAFSSDSYF-LASRN-D--NVPNA---VWIWD WD 9 361-401 IQKLKLFVVLEH--MS---PVRSFQWDPQQPR-LAICT-G--G--SK---VYLWS WD10 405-443 -CVS-VQVPGEG--DF---PVLGLCWHLSGDS-LALLS-K--D--HF---CLCFL WD40 Template TGEC-LRTLSGH--TS---GVTSVSFSPDGKR-LVSGS-E--D--GT---IKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |