WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000112282 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000112282 (Len=485) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000112282, Len=485): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 GFICSENGDESSSDESAQSPGAEKCCLASDLMKLCGETKARNKARRRTTTQMELLYAHSGDPDAPPEDFSSPLLPVVESP 80
WD40-rpts (________________________WD 1________________________) (_________________WD
Query 81 EGNKELTPAPYSGDREPVTKINAVQSEVNSSARSPTATERKSLDRSPLTRRRMQERSQNHTDRSRTLDKPGVASPALAAR 160
WD40-rpts 2________________)_______________WD 3_______________)(_______________WD 4_______
Query 161 SSRAARLQCVHVAEGHTKPLLCLDSTDDLLFTGSKDRTCKVWNLVTGQEIMSLGDHPSSVVSVRYCSSLVFTVSTAYVKV 240
WD40-rpts ________)______________WD 5_______________) (_______________WD 6______________
Query 241 WDIRDSAKCIRTLTSSGQVSQGDACGGSAVAIPPGENQINQIALNPSGTFLYAASGNTVRMWDLRRFVSTGKLTGHLGPV 320
WD40-rpts _) (________________WD 7________________) (_____________
Query 321 MCLTVDQMGKGQDVVITGSKDHTVKMFDVLEGAQGSISAAHHFDPAHQDGLEAVATQGDSLFTAARDSCIKKWDLASKQM 400
WD40-rpts _____WD 8__________________) (__________
Query 401 QEQVERAHCDPLSSLAVVPGSSAVVTGCKGGLLKFWHTHTLRALGEVRGHESSVNDVCTNSSQLFTASDDRTVKIWQVKG 480
WD40-rpts ___________WD 9_____________________) (________________WD10_______________)
Query 481 CLEDG 485
WD40-rpts
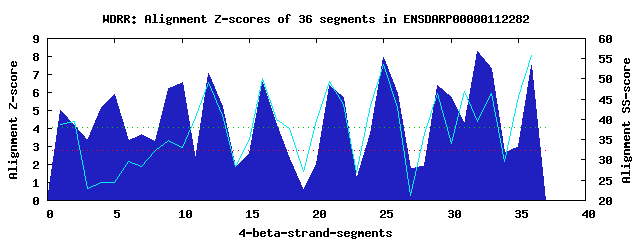 10 WD40-repeats found in QUERY=ENSDARP00000112282 (Len=485): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 3-56 5.00 321.21 0.00292 2.5% 38.88 WD 2: Seg 5 60-98 5.90 358.45 0.00092 15.0% 24.42 WD 3: Seg 7 97-133 3.69 266.91 0.01564 15.0% 28.50 WD 4: Seg 10 134-169 6.58 386.57 0.00038 5.0% 33.13 WD 5: Seg 12 168-203 7.05 405.75 0.00021 30.0% 49.08 WD 6: Seg 16 207-242 6.53 384.58 0.00041 32.5% 50.12 WD 7: Seg 21 266-303 6.41 379.65 0.00048 20.0% 49.27 WD 8: Seg 25 307-348 7.92 441.82 6.9e-05 30.0% 53.49 WD 9: Seg 32 390-437 8.27 456.10 4.4e-05 20.0% 39.47 WD10: Seg 36 441-477 7.50 424.52 0.00012 30.0% 55.90Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 3-56 ICSENGDESSSDESAQSPGAEKCC--------LASDLMKLCGETKAR---NKARRRTTTQMELLY WD 2 60-98 -G--------------DPDAPPED--------FSSPLLPVVESPEGN---KELTPAPYSGDREPV WD 3 97-133 -----------------PVTKINA--------VQSEVNSSARSPTAT---ERKSLDRSPLTRRRM WD 4 134-169 ------------------QERSQN--------HTDRSRTLDKPGVAS---PALAARSSRAARLQC WD 5 168-203 ----------------QCVHVAEG--------HTKPL--LCLDSTDD---LLFTGSKDRTCKVWN WD 6 207-242 -G--------------QEIMSLGD--------HPSSVVSVRYCSSLV---FTVS---TAYVKVWD WD 7 266-303 -G--------------GSAVAIPP--------GENQINQIALNPSGT---FLYAAS-GNTVRMWD WD 8 307-348 -F--------------VSTGKLTG--------HLGPVMCLTVDQMGKGQDVVITGSKDHTVKMFD WD 9 390-437 IK--------------KWDLASKQMQEQVERAHCDPLSSLAVVPGSS---AVVTGCKGGLLKFWH WD10 441-477 -L--------------RALGEVRG--------HESSVNDVCT--NSS---QLFTASDDRTVKIWQ WD40 Template TG--------------ECLRTLSG--------HTSGVTSVSFSPDGK---RLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |