WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000111837 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000111837 (Len=256) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000111837, Len=256): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MMGLDQDQVWIGSEDSVIYIINTRSMLCHKQLTEHRGELTDFTLEKLSPESRSSRAQVYSCSADGTVIVWDVPSLKVKRQ 80
WD40-rpts (____________________WD 1____________________) (_____
Query 81 FHLSCDRLQSIQIHNGALWCVCEGARDCIMMLKPNGDQLWKIHLPDKMKATANFFNRVTLFPERQQIWTSFTDFAELCIW 160
WD40-rpts ____________WD 2________________) (________________WD 3________________
Query 161 DFNDPVKPPKRLKLPNCKGVTCMIKVKKQVWIGCGGGSGLSKVVGKILVLDSESLEVEKELEAHEDSVQVLFSVEDRYVL 240
WD40-rpts ) (______________________WD 4________
Query 241 SGSAKLEGRIAIWRVD 256
WD40-rpts _____________)
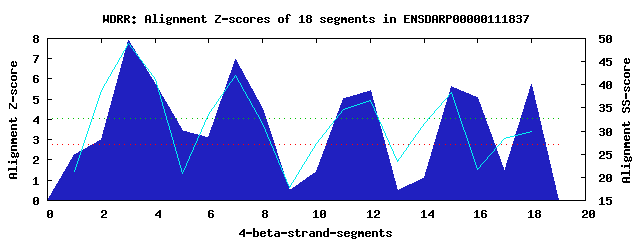 4 WD40-repeats found in QUERY=ENSDARP00000111837 (Len=256): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 3 26-71 7.88 440.30 7.2e-05 30.0% 48.98 WD 2: Seg 7 75-113 6.97 402.53 0.00023 10.0% 42.08 WD 3: Seg 12 124-161 5.41 338.01 0.00173 12.5% 36.58 WD 4: Seg 18 206-254 5.71 350.56 0.00117 22.5% 29.81Multiple sequence alignment of 4 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 26-71 -MLCHKQLT---------EHRGELTDFTLEKLSPESRSSRAQV-Y-SCSADGTVIVWD WD 2 75-113 -LKVKRQFH---------LSCDRLQSIQIHN-------GALWC-V-CEGARDCIMMLK WD 3 124-161 LPD---KMK---------ATANFFNRVTLFP-------ERQQIWT-SFTDFAELCIWD WD 4 206-254 KILVL-DSESLEVEKELEAHEDSVQVLFSVE-------DRYVL-SGSAKLEGRIAIWR WD40 Template TGECLRTLS---------GHTSGVTSVSFSP-------DGKRL-V-SGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |