WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000105980 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000105980 (Len=534) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000105980, Len=534): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MSGVAQSGEGAAGQEAEADGEPPHEALVQGALEEEEEPAAKQPRLQQEDVALDFPMIEIAHEPSEEKSKSQDAAGVEEEV 80
WD40-rpts (_________________WD 1_________________) (_________________WD 2________________
Query 81 SYHQTDEPVHVAEHAEMERDNEWHQNGAESREMLEQTEENTADEPEKNSPEAEQHYQGLDFRQNPQMLTGSWAEYTHSAE 160
WD40-rpts ) (__________________WD 3__________________) (__________
Query 161 NYLRGCKWAPDGSCIVSNSADNVLRVYNLPAELYSSQWDLLSEMSPVLKMAEGDTIYDYCWFPKMTSTDPDTCFIASSSR 240
WD40-rpts _______WD 4________________) (________________WD 5_________
Query 241 DNPVHIWDAFYGDLRASFRPYNHLDELTAAHSLCFSPDGSQLYCGFDKIVRVFHTDRPGRDCEQRPTMVKKRGQTGIISC 320
WD40-rpts _______) (___________________WD 6__________________) (_______________
Query 321 IAFSQCHSMYACGSYSRSVGLYSCDDGSLLALLPTRHHGGLTHLLFSPNGYHLYTGGRKDSEILCWDLRDPGQVLFSMQR 400
WD40-rpts __WD 7________________) (__________________WD 8_________________) (________
Query 401 NVNTNQRIYFDLDQSGRYLLSGDTDGVVSVWDTLTALPDGNEEILKPLLQFQAHTDCTNGISVHPFMPLMASSSGQRKFF 480
WD40-rpts __________WD 9_________________) (________________WD10_______________
Query 481 WPSDSEDSTSDSEEGNVMPTNGDRPDNALVLWWAGPVASTNEIRQEEEPAVIDS 534
WD40-rpts _)(_____________WD11____________)
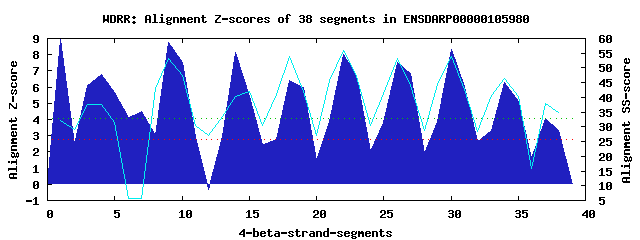 11 WD40-repeats found in QUERY=ENSDARP00000105980 (Len=534): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 1-40 8.95 484.22 1.9e-05 12.5% 32.05 WD 2: Seg 4 43-81 6.75 393.62 0.00031 7.5% 37.66 WD 3: Seg 7 85-126 4.48 299.55 0.00571 10.0% 5.79 WD 4: Seg 9 150-188 8.73 475.33 2.4e-05 22.5% 53.27 WD 5: Seg 14 211-248 8.13 450.49 5.3e-05 22.5% 40.23 WD 6: Seg 18 252-294 6.43 380.28 0.00047 27.5% 53.99 WD 7: Seg 22 305-343 8.03 446.55 6.0e-05 15.0% 55.77 WD 8: Seg 26 347-387 7.50 424.44 0.00012 40.0% 53.06 WD 9: Seg 30 392-432 8.31 457.91 4.2e-05 32.5% 53.96 WD10: Seg 34 445-482 6.27 373.84 0.00057 20.0% 46.34 WD11: Seg 38 483-513 3.34 252.65 0.02425 15.0% 34.95Multiple sequence alignment of 11 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 1-40 MSGVAQSG-EGAA-----GQEAEADGEPP--HEALV----QGA-L--EEEEEPAA WD 2 43-81 -PRLQQED-VALD-----FPMIEIAHEPS--EEKSK----SQD-A--AGVEEEVS WD 3 85-126 -TDEPVHV-AEHA-----EMERDNEWHQN--GAES-REMLEQT-E--ENTADEPE WD 4 150-188 -GSWAEYT-HSAE-----NYLRGCKWAPD--GSCIV----SNS-A--DNVLRVYN WD 5 211-248 --AEGDTI-YDYC-----WFPKMTSTDPD--TCFIA----SSS-R--DNPVHIWD WD 6 252-294 -GDLRASF-RPYNHLDELTAAHSLCFSPD--GSQLY----CGF----DKIVRVFH WD 7 305-343 -RPTMVKK-RGQT-----GIISCIAFSQC--HSMYA----CGS-Y--SRSVGLYS WD 8 347-387 -GSLLALLPTRHH-----GGLTHLLFSPN--GYHLY----TGGRK--DSEILCWD WD 9 392-432 -GQVLFSM-QRNV-----NTNQRIYFDLDQSGRYLL----SGD-T--DGVVSVWD WD10 445-482 -LKPLLQF-QAHT-----DCTNGISVHPF--MPLMA----SSS-G--QRKF-FWP WD11 483-513 ------------S-----DSEDSTSDSEE--GNVMP----TNG-DRPDNALVLWW WD40 Template TGECLRTL-SGHT-----SGVTSVSFSPD--GKRLV----SGS-E--DGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |